Oncostatin M inhibits myoblast differentiation and regulates muscle regeneration
- PMID: 20956996
- PMCID: PMC3193431
- DOI: 10.1038/cr.2010.144
Oncostatin M inhibits myoblast differentiation and regulates muscle regeneration
Abstract
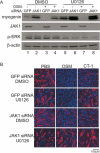
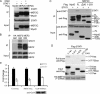
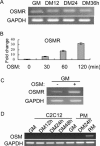
Oncostatin M (OSM) is a cytokine of the interleukin-6 family and plays important roles during inflammation. However, its roles in myoblast differentiation and muscle regeneration remain unexplored. We show here that OSM potently inhibited myoblast differentiation mainly by activating the JAK1/STAT1/STAT3 pathway. OSM downregulated myocyte enhancer-binding factor 2A (MEF2A), upregulated the expression of Id1 and Id2, and inhibited the transcriptional activity of MyoD and MEF2. In addition, OSM also enhanced the expression of STAT3 and OSM receptor, which constituted a positive feedback loop to further amplify OSM-induced signaling. Moreover, we found that STAT1 physically associated with MEF2 and repressed its transcriptional activity, which could account for the OSM-mediated repression of MEF2. Although undetectable in normal muscles in vivo, OSM was rapidly induced on muscle injury and then promptly downregulated just before the majority of myoblasts differentiate. Prolonged expression of OSM in muscles compromised the regeneration process without affecting myoblast proliferation, suggesting that OSM functions to prevent proliferating myoblasts from premature differentiation during the early phase of muscle regeneration.
Figures




Similar articles
-
JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts.J Cell Biol. 2007 Oct 8;179(1):129-38. doi: 10.1083/jcb.200703184. Epub 2007 Oct 1. J Cell Biol. 2007. PMID: 17908914 Free PMC article.
-
Oncostatin M induces C2C12 myotube atrophy by modulating muscle differentiation and degradation.Biochem Biophys Res Commun. 2019 Aug 27;516(3):951-956. doi: 10.1016/j.bbrc.2019.06.143. Epub 2019 Jul 2. Biochem Biophys Res Commun. 2019. PMID: 31272716
-
Role of JAK3 in myogenic differentiation.Cell Signal. 2012 Mar;24(3):742-9. doi: 10.1016/j.cellsig.2011.11.009. Epub 2011 Nov 18. Cell Signal. 2012. PMID: 22120524
-
MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation.Curr Opin Cell Biol. 1999 Dec;11(6):683-8. doi: 10.1016/s0955-0674(99)00036-8. Curr Opin Cell Biol. 1999. PMID: 10600704 Review.
-
Mef2 and the skeletal muscle differentiation program.Semin Cell Dev Biol. 2017 Dec;72:33-44. doi: 10.1016/j.semcdb.2017.11.020. Epub 2017 Nov 22. Semin Cell Dev Biol. 2017. PMID: 29154822 Review.
Cited by
-
Expression of nephronectin is inhibited by oncostatin M via both JAK/STAT and MAPK pathways.FEBS Open Bio. 2015 Apr 8;5:303-7. doi: 10.1016/j.fob.2015.04.001. eCollection 2015. FEBS Open Bio. 2015. PMID: 25905035 Free PMC article.
-
Emerging Mechanisms of Skeletal Muscle Homeostasis and Cachexia: The SUMO Perspective.Cells. 2023 Feb 17;12(4):644. doi: 10.3390/cells12040644. Cells. 2023. PMID: 36831310 Free PMC article. Review.
-
JAK-STAT pathway and myogenic differentiation.JAKSTAT. 2013 Apr 1;2(2):e23282. doi: 10.4161/jkst.23282. JAKSTAT. 2013. PMID: 24058805 Free PMC article. Review.
-
Exogenous Oncostatin M induces Cardiac Dysfunction, Musculoskeletal Atrophy, and Fibrosis.Cytokine. 2022 Nov;159:155972. doi: 10.1016/j.cyto.2022.155972. Epub 2022 Aug 30. Cytokine. 2022. PMID: 36054964 Free PMC article.
-
The Role of Oxidative Stress in Skeletal Muscle Myogenesis and Muscle Disease.Antioxidants (Basel). 2022 Apr 11;11(4):755. doi: 10.3390/antiox11040755. Antioxidants (Basel). 2022. PMID: 35453440 Free PMC article. Review.
References
-
- Buckingham M. Skeletal muscle formation in vertebrates. Curr Opin Genet Dev. 2001;11:440–448. - PubMed
-
- Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. - PubMed
-
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. - PubMed
-
- Arnold HH, Braun T. Targeted inactivation of myogenic factor genes reveals their role during mouse myogenesis: a review. Int J Dev Biol. 1996;40:345–353. - PubMed
-
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

