The mammalian Sin3 proteins are required for muscle development and sarcomere specification
- PMID: 20956564
- PMCID: PMC3004272
- DOI: 10.1128/MCB.00975-10
The mammalian Sin3 proteins are required for muscle development and sarcomere specification
Abstract
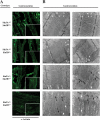
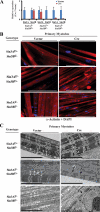
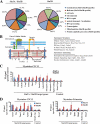
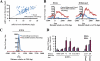
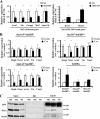
The highly related mammalian Sin3A and Sin3B proteins provide a versatile platform for chromatin-modifying activities. Sin3-containing complexes play a role in gene repression through deacetylation of nucleosomes. Here, we explore a role for Sin3 in myogenesis by examining the phenotypes resulting from acute somatic deletion of both isoforms in vivo and from primary myotubes in vitro. Myotubes ablated for Sin3A alone, but not Sin3B, displayed gross defects in sarcomere structure that were considerably enhanced upon simultaneous ablation of both isoforms. Massively parallel sequencing of Sin3A- and Sin3B-bound genomic loci revealed a subset of target genes directly involved in sarcomere function that are positively regulated by Sin3A and Sin3B proteins. Both proteins were coordinately recruited to a substantial number of genes. Interestingly, depletion of Sin3B led to compensatory increases in Sin3A recruitment at certain target loci, but Sin3B was never found to compensate for Sin3A loss. Thus, our analyses describe a novel transcriptional role for Sin3A and Sin3B proteins associated with maintenance of differentiated muscle cells.
Figures


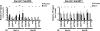
Similar articles
-
SIN3A and SIN3B differentially regulate breast cancer metastasis.Oncotarget. 2016 Nov 29;7(48):78713-78725. doi: 10.18632/oncotarget.12805. Oncotarget. 2016. PMID: 27780928 Free PMC article.
-
Haploinsufficiency of the Sin3/HDAC corepressor complex member SIN3B causes a syndromic intellectual disability/autism spectrum disorder.Am J Hum Genet. 2021 May 6;108(5):929-941. doi: 10.1016/j.ajhg.2021.03.017. Epub 2021 Apr 2. Am J Hum Genet. 2021. PMID: 33811806 Free PMC article.
-
Differential Complex Formation via Paralogs in the Human Sin3 Protein Interaction Network.Mol Cell Proteomics. 2020 Sep;19(9):1468-1484. doi: 10.1074/mcp.RA120.002078. Epub 2020 May 28. Mol Cell Proteomics. 2020. PMID: 32467258 Free PMC article.
-
Sin3: insight into its transcription regulatory functions.Eur J Cell Biol. 2013 Aug-Sep;92(8-9):237-46. doi: 10.1016/j.ejcb.2013.09.001. Epub 2013 Oct 9. Eur J Cell Biol. 2013. PMID: 24189169 Review.
-
Emerging Roles of Epigenetic Regulator Sin3 in Cancer.Adv Cancer Res. 2016;130:113-35. doi: 10.1016/bs.acr.2016.01.006. Epub 2016 Mar 2. Adv Cancer Res. 2016. PMID: 27037752 Review.
Cited by
-
The HDAC-Associated Sin3B Protein Represses DREAM Complex Targets and Cooperates with APC/C to Promote Quiescence.Cell Rep. 2018 Dec 4;25(10):2797-2807.e8. doi: 10.1016/j.celrep.2018.11.024. Cell Rep. 2018. PMID: 30517867 Free PMC article.
-
Transcriptional co-repressors and memory storage.Neuropharmacology. 2014 May;80:53-60. doi: 10.1016/j.neuropharm.2014.01.003. Epub 2014 Jan 17. Neuropharmacology. 2014. PMID: 24440532 Free PMC article. Review.
-
Coregulator Sin3a Promotes Postnatal Murine β-Cell Fitness by Regulating Genes in Ca2+ Homeostasis, Cell Survival, Vesicle Biosynthesis, Glucose Metabolism, and Stress Response.Diabetes. 2020 Jun;69(6):1219-1231. doi: 10.2337/db19-0721. Epub 2020 Apr 3. Diabetes. 2020. PMID: 32245798 Free PMC article.
-
Identification of long noncoding RNAs involved in muscle differentiation.PLoS One. 2018 Mar 2;13(3):e0193898. doi: 10.1371/journal.pone.0193898. eCollection 2018. PLoS One. 2018. PMID: 29499054 Free PMC article.
-
The potential of targeting Sin3B and its associated complexes for cancer therapy.Expert Opin Ther Targets. 2017 Nov;21(11):1051-1061. doi: 10.1080/14728222.2017.1386655. Epub 2017 Oct 9. Expert Opin Ther Targets. 2017. PMID: 28956957 Free PMC article. Review.
References
-
- Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. - PubMed
-
- Ayer, D. E., Q. A. Lawrence, and R. N. Eisenman. 1995. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell 80:767-776. - PubMed
-
- Batchelor, C. L., and S. J. Winder. 2006. Sparks, signals and shock absorbers: how dystrophin loss causes muscular dystrophy. Trends Cell Biol. 16:198-205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
