Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches
- PMID: 20942956
- PMCID: PMC3091713
- DOI: 10.1186/1471-2164-11-564
Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches
Abstract
Background: Rainbow trout are important fish for aquaculture and recreational fisheries and serves as a model species for research investigations associated with carcinogenesis, comparative immunology, toxicology and evolutionary biology. However, to date there is no genome reference sequence to facilitate the development of molecular technologies that utilize high-throughput characterizations of gene expression and genetic variation. Alternatively, transcriptome sequencing is a rapid and efficient means for gene discovery and genetic marker development. Although a large number (258,973) of EST sequences are publicly available, the nature of rainbow trout duplicated genome hinders assembly and complicates annotation.
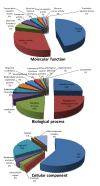
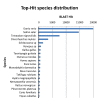
Results: High-throughput deep sequencing of the Swanson rainbow trout doubled-haploid transcriptome using 454-pyrosequencing technology yielded ~1.3 million reads with an average length of 344 bp, a total of 447 million bases. De novo assembly of the sequences yielded 151,847 Tentative Consensus (TC) sequences (average length of 662 bp) and 224,391 singletons. A combination assembly of both the 454-pyrosequencing ESTs and the pre-existing sequences resulted in 161,818 TCs (average length of 758 bp) and 261,071 singletons. Gene Ontology analysis of the combination assembly showed high similarities to transcriptomes of other fish species with known genome sequences.
Conclusion: The 454 library significantly increased the suite of ESTs available for rainbow trout, allowing improved assembly and annotation of the transcriptome. Furthermore, the 454 sequencing enables functional genome research in rainbow trout, providing a wealth of sequence data to serve as a reference transcriptome for future studies including identification of paralogous sequences and/or allelic variation, digital gene expression and proteomic research.
Figures
Similar articles
-
Transcriptome assembly, gene annotation and tissue gene expression atlas of the rainbow trout.PLoS One. 2015 Mar 20;10(3):e0121778. doi: 10.1371/journal.pone.0121778. eCollection 2015. PLoS One. 2015. PMID: 25793877 Free PMC article.
-
Single nucleotide polymorphism discovery in rainbow trout by deep sequencing of a reduced representation library.BMC Genomics. 2009 Nov 25;10:559. doi: 10.1186/1471-2164-10-559. BMC Genomics. 2009. PMID: 19939274 Free PMC article.
-
Analysis of BAC-end sequences in rainbow trout: content characterization and assessment of synteny between trout and other fish genomes.BMC Genomics. 2011 Jun 14;12:314. doi: 10.1186/1471-2164-12-314. BMC Genomics. 2011. PMID: 21672188 Free PMC article.
-
Status and opportunities for genomics research with rainbow trout.Comp Biochem Physiol B Biochem Mol Biol. 2002 Dec;133(4):609-46. doi: 10.1016/s1096-4959(02)00167-7. Comp Biochem Physiol B Biochem Mol Biol. 2002. PMID: 12470823 Review.
-
Next-generation transcriptome assembly.Nat Rev Genet. 2011 Sep 7;12(10):671-82. doi: 10.1038/nrg3068. Nat Rev Genet. 2011. PMID: 21897427 Review.
Cited by
-
Integrated application of transcriptomics and metabolomics yields insights into population-asynchronous ovary development in Coilia nasus.Sci Rep. 2016 Aug 22;6:31835. doi: 10.1038/srep31835. Sci Rep. 2016. PMID: 27545088 Free PMC article.
-
A synthetic rainbow trout linkage map provides new insights into the salmonid whole genome duplication and the conservation of synteny among teleosts.BMC Genet. 2012 Mar 16;13:15. doi: 10.1186/1471-2156-13-15. BMC Genet. 2012. PMID: 22424132 Free PMC article.
-
A second generation integrated map of the rainbow trout (Oncorhynchus mykiss) genome: analysis of conserved synteny with model fish genomes.Mar Biotechnol (NY). 2012 Jun;14(3):343-57. doi: 10.1007/s10126-011-9418-z. Epub 2011 Nov 19. Mar Biotechnol (NY). 2012. PMID: 22101344
-
Characterization of the rainbow trout spleen transcriptome and identification of immune-related genes.Front Genet. 2014 Oct 14;5:348. doi: 10.3389/fgene.2014.00348. eCollection 2014. Front Genet. 2014. PMID: 25352861 Free PMC article.
-
Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition).Autophagy. 2016;12(1):1-222. doi: 10.1080/15548627.2015.1100356. Autophagy. 2016. PMID: 26799652 Free PMC article. No abstract available.
References
-
- Harvey DJ. Aquaculture Outlook. Electronic Outlook Report from the Economic Research Service. 2006. http://www.ers.usda.gov
-
- Wolf K, Rumsey G. The representative research animal: why rainbow trout? Salmo gairdneri? Z Angew Ichthyol. 1985;3:131–138. doi: 10.1111/j.1439-0426.1985.tb00422.x. - DOI
-
- Behnke RJ. Native Trout of Western North America. American Fisheries Society Monograph 6, American Fisheries Society, Bethesda, MD. 1992.
-
- Thorgaard GH, Bailey GS, Williams D, Buhler DR, Kaattari SL, Ristow SS, Hansen JD, Winton JR, Bartholomew JL, Nagler JJ. et al.Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 2002;133(4):609–646. doi: 10.1016/S1096-4959(02)00167-7. - DOI - PubMed
-
- Davidson W, Chevalet C, Rexroad Iii CE, Omholt S. Salmonid Genomic Sequencing Initiative: the Case for Sequencing the Genomics of Atlantic Salmon (Salmo Salar) and Rainbow Trout (Oncorynchus Mykiss) Government Publication/report. 2006.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

