Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores
- PMID: 20937697
- PMCID: PMC2958477
- DOI: 10.1083/jcb.201006129
Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores
Abstract
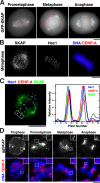
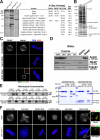
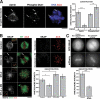
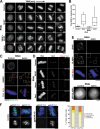
During mitosis, kinetochores play multiple roles to generate interactions with microtubules, and direct chromosome congression, biorientation, error correction, and anaphase segregation. However, it is unclear what changes at the kinetochore facilitate these distinct activities. Here, we describe a complex of the spindle- and kinetochore-associated protein Astrin, the small kinetochore-associated protein (SKAP), and the dynein light chain LC8. Although most dynein-associated proteins localize to unaligned kinetochores in an Aurora B-dependent manner, Astrin, SKAP, and LC8 localization is antagonized by Aurora B such that they target exclusively to bioriented kinetochores. Astrin-SKAP-depleted cells fail to maintain proper chromosome alignment, resulting in a spindle assembly checkpoint-dependent mitotic delay. Consistent with a role in stabilizing bioriented attachments, Astrin and SKAP bind directly to microtubules and are required for CLASP localization to kinetochores. In total, our results suggest that tension-dependent Aurora B phosphorylation can act to control outer kinetochore composition to provide distinct activities to prometaphase and metaphase kinetochores.
Figures

Similar articles
-
The Astrin-SKAP complex reduces friction at the kinetochore-microtubule interface.Curr Biol. 2022 Jun 20;32(12):2621-2631.e3. doi: 10.1016/j.cub.2022.04.061. Epub 2022 May 16. Curr Biol. 2022. PMID: 35580605 Free PMC article.
-
Aurora B kinase-dependent recruitment of hZW10 and hROD to tensionless kinetochores.Curr Biol. 2007 Dec 18;17(24):2143-9. doi: 10.1016/j.cub.2007.11.037. Epub 2007 Dec 6. Curr Biol. 2007. PMID: 18065224
-
Zwint-1 is a novel Aurora B substrate required for the assembly of a dynein-binding platform on kinetochores.Mol Biol Cell. 2011 Sep;22(18):3318-30. doi: 10.1091/mbc.E11-03-0213. Epub 2011 Jul 20. Mol Biol Cell. 2011. PMID: 21775627 Free PMC article.
-
Regulation of kinetochore-microtubule attachments by Aurora B kinase.Biochem Soc Trans. 2009 Oct;37(Pt 5):976-80. doi: 10.1042/BST0370976. Biochem Soc Trans. 2009. PMID: 19754435 Review.
-
Killing two birds with one stone: how budding yeast Mps1 controls chromosome segregation and spindle assembly checkpoint through phosphorylation of a single kinetochore protein.Curr Genet. 2020 Dec;66(6):1037-1044. doi: 10.1007/s00294-020-01091-x. Epub 2020 Jul 6. Curr Genet. 2020. PMID: 32632756 Review.
Cited by
-
Signaling Scaffold Protein IQGAP1 Interacts with Microtubule Plus-end Tracking Protein SKAP and Links Dynamic Microtubule Plus-end to Steer Cell Migration.J Biol Chem. 2015 Sep 25;290(39):23766-80. doi: 10.1074/jbc.M115.673517. Epub 2015 Aug 4. J Biol Chem. 2015. PMID: 26242911 Free PMC article.
-
The lethal response to Cdk1 inhibition depends on sister chromatid alignment errors generated by KIF4 and isoform 1 of PRC1.Sci Rep. 2015 Oct 1;5:14798. doi: 10.1038/srep14798. Sci Rep. 2015. PMID: 26423135 Free PMC article.
-
Structural features of LC8-induced self-association of swallow.Biochemistry. 2013 Sep 3;52(35):6011-20. doi: 10.1021/bi400642u. Epub 2013 Aug 21. Biochemistry. 2013. PMID: 23914803 Free PMC article.
-
Recent Research Advances in Mitosis during Mammalian Gametogenesis.Cells. 2019 Jun 10;8(6):567. doi: 10.3390/cells8060567. Cells. 2019. PMID: 31185583 Free PMC article. Review.
-
Dynein light intermediate chains maintain spindle bipolarity by functioning in centriole cohesion.J Cell Biol. 2014 Nov 24;207(4):499-516. doi: 10.1083/jcb.201408025. J Cell Biol. 2014. PMID: 25422374 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

