Next-generation sequencing identifies the natural killer cell microRNA transcriptome
- PMID: 20935160
- PMCID: PMC2963822
- DOI: 10.1101/gr.107995.110
Next-generation sequencing identifies the natural killer cell microRNA transcriptome
Abstract
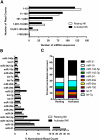
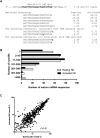
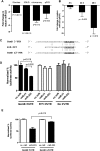
Natural killer (NK) cells are innate lymphocytes important for early host defense against infectious pathogens and surveillance against malignant transformation. Resting murine NK cells regulate the translation of effector molecule mRNAs (e.g., granzyme B, GzmB) through unclear molecular mechanisms. MicroRNAs (miRNAs) are small noncoding RNAs that post-transcriptionally regulate the translation of their mRNA targets, and are therefore candidates for mediating this control process. While the expression and importance of miRNAs in T and B lymphocytes have been established, little is known about miRNAs in NK cells. Here, we used two next-generation sequencing (NGS) platforms to define the miRNA transcriptomes of resting and cytokine-activated primary murine NK cells, with confirmation by quantitative real-time PCR (qRT-PCR) and microarrays. We delineate a bioinformatics analysis pipeline that identified 302 known and 21 novel mature miRNAs from sequences obtained from NK cell small RNA libraries. These miRNAs are expressed over a broad range and exhibit isomiR complexity, and a subset is differentially expressed following cytokine activation. Using these miRNA NGS data, miR-223 was identified as a mature miRNA present in resting NK cells with decreased expression following cytokine activation. Furthermore, we demonstrate that miR-223 specifically targets the 3' untranslated region of murine GzmB in vitro, indicating that this miRNA may contribute to control of GzmB translation in resting NK cells. Thus, the sequenced NK cell miRNA transcriptome provides a valuable framework for further elucidation of miRNA expression and function in NK cell biology.
Figures


Similar articles
-
Identification of microRNA transcriptome involved in human natural killer cell activation.Immunol Lett. 2012 Apr 30;143(2):208-17. doi: 10.1016/j.imlet.2012.02.014. Immunol Lett. 2012. PMID: 22701882
-
MicroRNA Expression Analysis: Next-Generation Sequencing.Methods Mol Biol. 2018;1783:171-183. doi: 10.1007/978-1-4939-7834-2_8. Methods Mol Biol. 2018. PMID: 29767362
-
MicroRNA-deficient NK cells exhibit decreased survival but enhanced function.J Immunol. 2012 Apr 1;188(7):3019-30. doi: 10.4049/jimmunol.1102294. Epub 2012 Feb 29. J Immunol. 2012. PMID: 22379033 Free PMC article.
-
Computational Resources for Prediction and Analysis of Functional miRNA and Their Targetome.Methods Mol Biol. 2019;1912:215-250. doi: 10.1007/978-1-4939-8982-9_9. Methods Mol Biol. 2019. PMID: 30635896 Review.
-
microRNA management of NK-cell developmental and functional programs.Eur J Immunol. 2014 Oct;44(10):2862-8. doi: 10.1002/eji.201444798. Epub 2014 Sep 16. Eur J Immunol. 2014. PMID: 25142111 Free PMC article. Review.
Cited by
-
Overexpression of miR-155 causes expansion, arrest in terminal differentiation and functional activation of mouse natural killer cells.Blood. 2013 Apr 18;121(16):3126-34. doi: 10.1182/blood-2012-12-467597. Epub 2013 Feb 19. Blood. 2013. PMID: 23422749 Free PMC article.
-
Molecular mechanisms influencing NK cell development: implications for NK cell malignancies.Am J Blood Res. 2011;1(1):34-45. Epub 2011 May 22. Am J Blood Res. 2011. PMID: 22432064 Free PMC article.
-
MicroRNAs: Pleiotropic Regulators in the Tumor Microenvironment.Front Immunol. 2018 Nov 1;9:2491. doi: 10.3389/fimmu.2018.02491. eCollection 2018. Front Immunol. 2018. PMID: 30443251 Free PMC article. Review.
-
Stage-specific regulation of natural killer cell homeostasis and response against viral infection by microRNA-155.Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):6967-72. doi: 10.1073/pnas.1304410110. Epub 2013 Apr 9. Proc Natl Acad Sci U S A. 2013. PMID: 23572582 Free PMC article.
-
Comprehensive analysis of microRNAs in breast cancer.BMC Genomics. 2012;13 Suppl 7(Suppl 7):S18. doi: 10.1186/1471-2164-13-S7-S18. Epub 2012 Dec 13. BMC Genomics. 2012. PMID: 23281739 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
