Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM)
- PMID: 20930118
- PMCID: PMC2972957
- DOI: 10.1073/pnas.1008924107
Mitochondrial Lon protease regulates mitochondrial DNA copy number and transcription by selective degradation of mitochondrial transcription factor A (TFAM)
Abstract
Lon is the major protease in the mitochondrial matrix in eukaryotes, and is well conserved among species. Although a role for Lon in mitochondrial biogenesis has been proposed, the mechanistic basis is unclear. Here, we demonstrate a role for Lon in mtDNA metabolism. An RNA interference (RNAi) construct was designed that reduces Lon to less than 10% of its normal level in Drosophila Schneider cells. RNAi knockdown of Lon results in increased abundance of mitochondrial transcription factor A (TFAM) and mtDNA copy number. In a corollary manner, overexpression of Lon reduces TFAM levels and mtDNA copy number. Notably, induction of mtDNA depletion in Lon knockdown cells does not result in degradation of TFAM, thereby causing a dramatic increase in the TFAMmtDNA ratio. The increased TFAMmtDNA ratio in turn causes inhibition of mitochondrial transcription. We conclude that Lon regulates mitochondrial transcription by stabilizing the mitochondrial TFAMmtDNA ratio via selective degradation of TFAM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
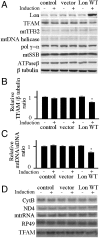
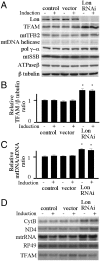
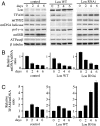

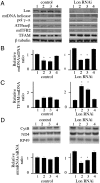
Similar articles
-
Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease.Mol Cell. 2013 Jan 10;49(1):121-32. doi: 10.1016/j.molcel.2012.10.023. Epub 2012 Nov 29. Mol Cell. 2013. PMID: 23201127 Free PMC article.
-
Drosophila mitochondrial transcription factor B2 regulates mitochondrial DNA copy number and transcription in schneider cells.J Biol Chem. 2004 Jun 25;279(26):26900-5. doi: 10.1074/jbc.M401643200. Epub 2004 Apr 1. J Biol Chem. 2004. PMID: 15060065
-
Tetramethylpyrazine blocks TFAM degradation and up-regulates mitochondrial DNA copy number by interacting with TFAM.Biosci Rep. 2017 May 17;37(3):BSR20170319. doi: 10.1042/BSR20170319. Print 2017 Jun 30. Biosci Rep. 2017. PMID: 28465355 Free PMC article.
-
Matrix proteases in mitochondrial DNA function.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):1080-7. doi: 10.1016/j.bbagrm.2011.11.008. Epub 2011 Dec 8. Biochim Biophys Acta. 2012. PMID: 22172992 Free PMC article. Review.
-
Mitochondrial transcription factor A regulates mitochondrial transcription initiation, DNA packaging, and genome copy number.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):921-9. doi: 10.1016/j.bbagrm.2012.03.002. Epub 2012 Mar 21. Biochim Biophys Acta. 2012. PMID: 22465614 Review.
Cited by
-
DNA polymerase gamma mutations that impair holoenzyme stability cause catalytic subunit depletion.Nucleic Acids Res. 2021 May 21;49(9):5230-5248. doi: 10.1093/nar/gkab282. Nucleic Acids Res. 2021. PMID: 33956154 Free PMC article.
-
Resveratrol enhances exercise training responses in rats selectively bred for high running performance.Food Chem Toxicol. 2013 Nov;61:53-9. doi: 10.1016/j.fct.2013.01.051. Epub 2013 Feb 17. Food Chem Toxicol. 2013. PMID: 23422033 Free PMC article.
-
The manner in which DNA is packaged with TFAM has an impact on transcription activation and inhibition.FEBS Open Bio. 2012 Jun 12;2:145-50. doi: 10.1016/j.fob.2012.06.001. Print 2012. FEBS Open Bio. 2012. PMID: 23650593 Free PMC article.
-
CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease.Am J Hum Genet. 2015 Jan 8;96(1):121-35. doi: 10.1016/j.ajhg.2014.12.003. Am J Hum Genet. 2015. PMID: 25574826 Free PMC article.
-
Overexpression of Lon contributes to survival and aggressive phenotype of cancer cells through mitochondrial complex I-mediated generation of reactive oxygen species.Cell Death Dis. 2013 Jun 20;4(6):e681. doi: 10.1038/cddis.2013.204. Cell Death Dis. 2013. PMID: 23788038 Free PMC article.
References
-
- Koppen M, Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit Rev Biochem Mol Biol. 2007;42:221–242. - PubMed
-
- Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res Microbiol. 2006;157:701–713. - PubMed
-
- Chandu D, Nandi D. Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res Microbiol. 2004;155:710–719. - PubMed
-
- Park SC, et al. Oligomeric structure of the ATP-dependent protease La (Lon) of Escherichia coli. Mol Cells. 2006;21:129–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

