Inference of homologous recombination in bacteria using whole-genome sequences
- PMID: 20923983
- PMCID: PMC2998322
- DOI: 10.1534/genetics.110.120121
Inference of homologous recombination in bacteria using whole-genome sequences
Abstract
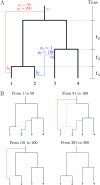
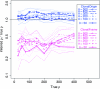
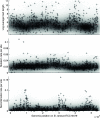
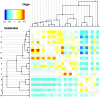
Bacteria and archaea reproduce clonally, but sporadically import DNA into their chromosomes from other organisms. In many of these events, the imported DNA replaces an homologous segment in the recipient genome. Here we present a new method to reconstruct the history of recombination events that affected a given sample of bacterial genomes. We introduce a mathematical model that represents both the donor and the recipient of each DNA import as an ancestor of the genomes in the sample. The model represents a simplification of the previously described coalescent with gene conversion. We implement a Monte Carlo Markov chain algorithm to perform inference under this model from sequence data alignments and show that inference is feasible for whole-genome alignments through parallelization. Using simulated data, we demonstrate accurate and reliable identification of individual recombination events and global recombination rate parameters. We applied our approach to an alignment of 13 whole genomes from the Bacillus cereus group. We find, as expected from laboratory experiments, that the recombination rate is higher between closely related organisms and also that the genome contains several broad regions of elevated levels of recombination. Application of the method to the genomic data sets that are becoming available should reveal the evolutionary history and private lives of populations of bacteria and archaea. The methods described in this article have been implemented in a computer software package, ClonalOrigin, which is freely available from http://code.google.com/p/clonalorigin/.
Figures


Similar articles
-
Inference of the properties of the recombination process from whole bacterial genomes.Genetics. 2014 Jan;196(1):253-65. doi: 10.1534/genetics.113.157172. Epub 2013 Oct 30. Genetics. 2014. PMID: 24172133 Free PMC article.
-
The Bacterial Sequential Markov Coalescent.Genetics. 2017 May;206(1):333-343. doi: 10.1534/genetics.116.198796. Epub 2017 Mar 3. Genetics. 2017. PMID: 28258183 Free PMC article.
-
ClonalFrameML: efficient inference of recombination in whole bacterial genomes.PLoS Comput Biol. 2015 Feb 12;11(2):e1004041. doi: 10.1371/journal.pcbi.1004041. eCollection 2015 Feb. PLoS Comput Biol. 2015. PMID: 25675341 Free PMC article.
-
Genomics of the Bacillus cereus group of organisms.FEMS Microbiol Rev. 2005 Apr;29(2):303-29. doi: 10.1016/j.femsre.2004.12.005. FEMS Microbiol Rev. 2005. PMID: 15808746 Review.
-
Unraveling recombination rate evolution using ancestral recombination maps.Bioessays. 2014 Sep;36(9):892-900. doi: 10.1002/bies.201400047. Epub 2014 Jul 14. Bioessays. 2014. PMID: 25043668 Free PMC article. Review.
Cited by
-
Patterns of gene flow define species of thermophilic Archaea.PLoS Biol. 2012 Feb;10(2):e1001265. doi: 10.1371/journal.pbio.1001265. Epub 2012 Feb 21. PLoS Biol. 2012. PMID: 22363207 Free PMC article.
-
Distinguishing imported cases from locally acquired cases within a geographically limited genomic sample of an infectious disease.Bioinformatics. 2023 Jan 1;39(1):btac761. doi: 10.1093/bioinformatics/btac761. Bioinformatics. 2023. PMID: 36440957 Free PMC article.
-
Recombination-aware phylogenetic analysis sheds light on the evolutionary origin of SARS-CoV-2.Sci Rep. 2024 Jan 4;14(1):541. doi: 10.1038/s41598-023-50952-1. Sci Rep. 2024. PMID: 38177346 Free PMC article.
-
A Bayesian approach to infer recombination patterns in coronaviruses.Nat Commun. 2022 Jul 20;13(1):4186. doi: 10.1038/s41467-022-31749-8. Nat Commun. 2022. PMID: 35859071 Free PMC article.
-
Coalescent framework for prokaryotes undergoing interspecific homologous recombination.Heredity (Edinb). 2018 May;120(5):474-484. doi: 10.1038/s41437-017-0034-1. Epub 2018 Jan 23. Heredity (Edinb). 2018. PMID: 29358726 Free PMC article.
References
-
- Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli et al., 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32 459–470. - PubMed
-
- Canchaya, C., G. Fournous, S. Chibani-Chennoufi, M. L. Dillmann and H. Brüssow, 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6 417–424. - PubMed
-
- Claverys, J. P., B. Martin and P. Polard, 2009. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 33 643–656. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

