Dynamics connect substrate recognition to catalysis in protein kinase A
- PMID: 20890288
- PMCID: PMC3487389
- DOI: 10.1038/nchembio.452
Dynamics connect substrate recognition to catalysis in protein kinase A
Erratum in
- Nat Chem Biol. 2011 May;7(5):319
Abstract
Atomic resolution studies of protein kinases have traditionally been carried out in the inhibitory state, limiting our current knowledge on the mechanisms of substrate recognition and catalysis. Using NMR, X-ray crystallography and thermodynamic measurements, we analyzed the substrate recognition process of cAMP-dependent protein kinase (PKA), finding that entropy and protein dynamics play a prominent role. The nucleotide acts as a dynamic and allosteric activator by coupling the two lobes of apo PKA, enhancing the enzyme dynamics synchronously and priming it for catalysis. The formation of the ternary complex is entropically driven, and NMR spin relaxation data reveal that both substrate and PKA are dynamic in the closed state. Our results show that the enzyme toggles between open and closed states, which indicates that a conformational selection rather than an induced-fit mechanism governs substrate recognition.
Figures
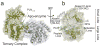


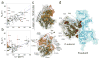
Similar articles
-
The αC-β4 loop controls the allosteric cooperativity between nucleotide and substrate in the catalytic subunit of protein kinase A.Elife. 2024 Jun 24;12:RP91506. doi: 10.7554/eLife.91506. Elife. 2024. PMID: 38913408 Free PMC article.
-
Allosteric cooperativity in protein kinase A.Proc Natl Acad Sci U S A. 2008 Jan 15;105(2):506-11. doi: 10.1073/pnas.0709214104. Epub 2008 Jan 4. Proc Natl Acad Sci U S A. 2008. PMID: 18178622 Free PMC article.
-
Mutation of a kinase allosteric node uncouples dynamics linked to phosphotransfer.Proc Natl Acad Sci U S A. 2017 Feb 7;114(6):E931-E940. doi: 10.1073/pnas.1620667114. Epub 2017 Jan 23. Proc Natl Acad Sci U S A. 2017. PMID: 28115705 Free PMC article.
-
Role of conformational entropy in the activity and regulation of the catalytic subunit of protein kinase A.FEBS J. 2013 Nov;280(22):5608-15. doi: 10.1111/febs.12462. Epub 2013 Aug 27. FEBS J. 2013. PMID: 23902454 Free PMC article. Review.
-
An NMR portrait of functional and dysfunctional allosteric cooperativity in cAMP-dependent protein kinase A.FEBS Lett. 2023 Apr;597(8):1055-1072. doi: 10.1002/1873-3468.14610. Epub 2023 Mar 26. FEBS Lett. 2023. PMID: 36892429 Free PMC article. Review.
Cited by
-
Heteronuclear Adiabatic Relaxation Dispersion (HARD) for quantitative analysis of conformational dynamics in proteins.J Magn Reson. 2012 Jun;219:75-82. doi: 10.1016/j.jmr.2012.03.024. Epub 2012 Apr 6. J Magn Reson. 2012. PMID: 22621977 Free PMC article.
-
Expanding the conformational selection paradigm in protein-ligand docking.Methods Mol Biol. 2012;819:59-74. doi: 10.1007/978-1-61779-465-0_5. Methods Mol Biol. 2012. PMID: 22183530 Free PMC article.
-
Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.Elife. 2022 Mar 17;11:e75346. doi: 10.7554/eLife.75346. Elife. 2022. PMID: 35297759 Free PMC article.
-
ATP-competitive inhibitors modulate the substrate binding cooperativity of a kinase by altering its conformational entropy.Sci Adv. 2022 Jul 29;8(30):eabo0696. doi: 10.1126/sciadv.abo0696. Epub 2022 Jul 29. Sci Adv. 2022. PMID: 35905186 Free PMC article.
-
Raf Kinase Inhibitory Protein regulates the cAMP-dependent protein kinase signaling pathway through a positive feedback loop.Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2121867119. doi: 10.1073/pnas.2121867119. Epub 2022 Jun 13. Proc Natl Acad Sci U S A. 2022. PMID: 35696587 Free PMC article.
References
-
- Walsh DA, Van Patten SM. Multiple pathway signal transduction by the cAMP-dependent protein kinase. FASEB J. 1994;8:1227–1236. - PubMed
-
- Shabb JB. Physiological substrates of cAMP-dependent protein kinase. Chem Rev. 2001;101:2381–2411. - PubMed
-
- Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, Anand G. PKA: A portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697:259–269. - PubMed
-
- Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101:2243–2270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR002301/RR/NCRR NIH HHS/United States
- HL080081/HL/NHLBI NIH HHS/United States
- P41 GM066326/GM/NIGMS NIH HHS/United States
- R01 GM019301/GM/NIGMS NIH HHS/United States
- GM072701/GM/NIGMS NIH HHS/United States
- P41RR02301/RR/NCRR NIH HHS/United States
- R01 GM072701-04/GM/NIGMS NIH HHS/United States
- P41GM66326/GM/NIGMS NIH HHS/United States
- R01 GM072701/GM/NIGMS NIH HHS/United States
- S10 RR008438/RR/NCRR NIH HHS/United States
- GM19301/GM/NIGMS NIH HHS/United States
- R37 GM019301/GM/NIGMS NIH HHS/United States
- R01 GM064742/GM/NIGMS NIH HHS/United States
- S10 RR002781/RR/NCRR NIH HHS/United States
- K02 HL080081/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

