Sequence requirements for ribosome stalling by the arginine attenuator peptide
- PMID: 20884617
- PMCID: PMC3003393
- DOI: 10.1074/jbc.M110.164152
Sequence requirements for ribosome stalling by the arginine attenuator peptide
Abstract
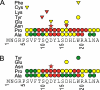
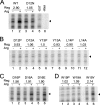
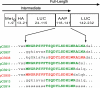
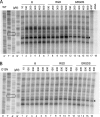
The 5' regions of eukaryotic mRNAs often contain upstream open reading frames (uORFs). The Neurospora crassa arg-2 uORF encodes the 24-residue arginine attenuator peptide (AAP). This regulatory uORF-encoded peptide, which is evolutionarily conserved in fungal transcripts specifying an arginine biosynthetic enzyme, functions as a nascent peptide within the ribosomal tunnel and negatively regulates gene expression. The nascent AAP causes ribosomes to stall at the uORF stop codon in response to arginine, thus, blocking ribosomes from reaching the ARG-2 initiation codon. Here scanning mutagenesis with alanine and proline was performed to systematically determine which AAP residues were important for conferring regulation. Changing many of the most highly conserved residues (Asp-12, Tyr-13, Lys-14, and Trp-19) abolished regulatory function. The minimal functional domain of the AAP was determined by positioning AAP sequences internally within a large polypeptide. Pulse-chase analyses revealed that residues 9-20 of the AAP composed the minimal domain that was sufficient to confer regulatory function. An extensive analysis of predicted fungal AAPs revealed that the minimal functional domain of the N. crassa AAP corresponded closely to the region that was most highly conserved among the fungi. We also observed that the tripeptide RGD could function similarly to arginine in triggering AAP-mediated ribosome stalling. These studies provide a better understanding of the elements required for a nascent peptide and a small regulatory molecule to control translational processes.
Figures



Similar articles
-
The arginine attenuator peptide interferes with the ribosome peptidyl transferase center.Mol Cell Biol. 2012 Jul;32(13):2396-406. doi: 10.1128/MCB.00136-12. Epub 2012 Apr 16. Mol Cell Biol. 2012. PMID: 22508989 Free PMC article.
-
The evolutionarily conserved eukaryotic arginine attenuator peptide regulates the movement of ribosomes that have translated it.Mol Cell Biol. 1998 Dec;18(12):7528-36. doi: 10.1128/MCB.18.12.7528. Mol Cell Biol. 1998. PMID: 9819438 Free PMC article.
-
Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation.J Biol Chem. 2000 Sep 1;275(35):26710-9. doi: 10.1074/jbc.M003175200. J Biol Chem. 2000. PMID: 10818103
-
Ribosome regulation by the nascent peptide.Microbiol Rev. 1996 Jun;60(2):366-85. doi: 10.1128/mr.60.2.366-385.1996. Microbiol Rev. 1996. PMID: 8801438 Free PMC article. Review.
-
Conserved Upstream Open Reading Frame Nascent Peptides That Control Translation.Annu Rev Genet. 2020 Nov 23;54:237-264. doi: 10.1146/annurev-genet-112618-043822. Epub 2020 Sep 1. Annu Rev Genet. 2020. PMID: 32870728 Free PMC article. Review.
Cited by
-
hnRNPs and ELAVL1 cooperate with uORFs to inhibit protein translation.Nucleic Acids Res. 2017 Mar;45(5):2849-2864. doi: 10.1093/nar/gkw991. Epub 2016 Oct 26. Nucleic Acids Res. 2017. PMID: 27789685 Free PMC article.
-
S-adenosyl-L-methionine induces compaction of nascent peptide chain inside the ribosomal exit tunnel upon translation arrest in the Arabidopsis CGS1 gene.J Biol Chem. 2011 Apr 29;286(17):14903-12. doi: 10.1074/jbc.M110.211656. Epub 2011 Feb 18. J Biol Chem. 2011. PMID: 21335553 Free PMC article.
-
Therapeutic targeting of YY1/MZF1 axis by MZF1-uPEP inhibits aerobic glycolysis and neuroblastoma progression.Theranostics. 2020 Jan 1;10(4):1555-1571. doi: 10.7150/thno.37383. eCollection 2020. Theranostics. 2020. PMID: 32042322 Free PMC article.
-
The role of upstream open reading frames in translation regulation in the apicomplexan parasites Plasmodium falciparum and Toxoplasma gondii.Parasitology. 2021 Sep;148(11):1277-1287. doi: 10.1017/S0031182021000937. Epub 2021 Jun 8. Parasitology. 2021. PMID: 34099078 Free PMC article. Review.
-
Crucial elements that maintain the interactions between the regulatory TnaC peptide and the ribosome exit tunnel responsible for Trp inhibition of ribosome function.Nucleic Acids Res. 2012 Mar;40(5):2247-57. doi: 10.1093/nar/gkr1052. Epub 2011 Nov 21. Nucleic Acids Res. 2012. PMID: 22110039 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

