Elicitation of structure-specific antibodies by epitope scaffolds
- PMID: 20876137
- PMCID: PMC2964213
- DOI: 10.1073/pnas.1004728107
Elicitation of structure-specific antibodies by epitope scaffolds
Abstract
Elicitation of antibodies against targets that are immunorecessive, cryptic, or transient in their native context has been a challenge for vaccine design. Here we demonstrate the elicitation of structure-specific antibodies against the HIV-1 gp41 epitope of the broadly neutralizing antibody 2F5. This conformationally flexible region of gp41 assumes mostly helical conformations but adopts a kinked, extended structure when bound by antibody 2F5. Computational techniques were employed to transplant the 2F5 epitope into select acceptor scaffolds. The resultant "2F5-epitope scaffolds" possessed nanomolar affinity for antibody 2F5 and a range of epitope flexibilities and antigenic specificities. Crystallographic characterization of the epitope scaffold with highest affinity and antigenic discrimination confirmed good to near perfect attainment of the target conformation for the gp41 molecular graft in free and 2F5-bound states, respectively. Animals immunized with 2F5-epitope scaffolds showed levels of graft-specific immune responses that correlated with graft flexibility (p < 0.04), while antibody responses against the graft-as dissected residue-by-residue with alanine substitutions-resembled more closely those of 2F5 than sera elicited with flexible or cyclized peptides, a resemblance heightened by heterologous prime-boost. Lastly, crystal structures of a gp41 peptide in complex with monoclonal antibodies elicited by the 2F5-epitope scaffolds revealed that the elicited antibodies induce gp41 to assume its 2F5-recognized shape. Epitope scaffolds thus provide a means to elicit antibodies that recognize a predetermined target shape and sequence, even if that shape is transient in nature, and a means by which to dissect factors influencing such elicitation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
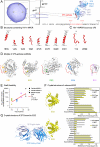

 , where i is the residue position at which the MPER was mutated to alanine. Alum/CpG, linked T help, increasing number of immunizations, heterologous immunizations, and use of ES5 all biased toward reduced R-values (
, where i is the residue position at which the MPER was mutated to alanine. Alum/CpG, linked T help, increasing number of immunizations, heterologous immunizations, and use of ES5 all biased toward reduced R-values (
Comment in
-
Scaffolding to build a rational vaccine design strategy.Proc Natl Acad Sci U S A. 2010 Oct 19;107(42):17859-60. doi: 10.1073/pnas.1012923107. Epub 2010 Oct 11. Proc Natl Acad Sci U S A. 2010. PMID: 20937874 Free PMC article. No abstract available.
Similar articles
-
Heterologous epitope-scaffold prime:boosting immuno-focuses B cell responses to the HIV-1 gp41 2F5 neutralization determinant.PLoS One. 2011 Jan 26;6(1):e16074. doi: 10.1371/journal.pone.0016074. PLoS One. 2011. PMID: 21297864 Free PMC article.
-
Structure-affinity relationships in the gp41 ELDKWA epitope for the HIV-1 neutralizing monoclonal antibody 2F5: effects of side-chain and backbone modifications and conformational constraints.J Pept Res. 2002 Jun;59(6):264-76. doi: 10.1034/j.1399-3011.2002.02988.x. J Pept Res. 2002. PMID: 12010517
-
Interactions of HIV-1 antibodies 2F5 and 4E10 with a gp41 epitope prebound to host and viral membrane model systems.Chembiochem. 2009 Apr 17;10(6):1032-44. doi: 10.1002/cbic.200800609. Chembiochem. 2009. PMID: 19283693
-
Progress towards the development of a HIV-1 gp41-directed vaccine.Curr HIV Res. 2004 Apr;2(2):193-204. doi: 10.2174/1570162043484933. Curr HIV Res. 2004. PMID: 15078183 Review.
-
Antigp41 membrane proximal external region antibodies and the art of using the membrane for neutralization.Curr Opin HIV AIDS. 2017 May;12(3):250-256. doi: 10.1097/COH.0000000000000364. Curr Opin HIV AIDS. 2017. PMID: 28422789 Review.
Cited by
-
Forced virus evolution reveals functional crosstalk between the disulfide bonded region and membrane proximal ectodomain region of HIV-1 gp41.Retrovirology. 2013 Apr 23;10:44. doi: 10.1186/1742-4690-10-44. Retrovirology. 2013. PMID: 23618462 Free PMC article.
-
Adding energy minimization strategy to peptide-design algorithm enables better search for RNA-binding peptides: Redesigned λ N peptide binds boxB RNA.J Comput Chem. 2016 Oct 15;37(27):2423-35. doi: 10.1002/jcc.24466. Epub 2016 Aug 4. J Comput Chem. 2016. PMID: 27487990 Free PMC article.
-
Scaffolding to build a rational vaccine design strategy.Proc Natl Acad Sci U S A. 2010 Oct 19;107(42):17859-60. doi: 10.1073/pnas.1012923107. Epub 2010 Oct 11. Proc Natl Acad Sci U S A. 2010. PMID: 20937874 Free PMC article. No abstract available.
-
Engineering, expression, purification, and characterization of stable clade A/B recombinant soluble heterotrimeric gp140 proteins.J Virol. 2012 Jan;86(1):128-42. doi: 10.1128/JVI.06363-11. Epub 2011 Oct 26. J Virol. 2012. PMID: 22031951 Free PMC article.
-
Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.Science. 2013 Nov 1;342(6158):592-8. doi: 10.1126/science.1243283. Science. 2013. PMID: 24179220 Free PMC article.
References
-
- Glennie MJ, Johnson PW. Clinical trials of antibody therapy. Immunol Today. 2000;21(8):403–410. - PubMed
-
- Fernandez-Carneado J, et al. Surface grafting onto template-assembled synthetic protein scaffolds in molecular recognition. Biopolymers. 2000;55(6):451–458. - PubMed
-
- Huang CC, et al. Scorpion-toxin mimics of CD4 in complex with human immunodeficiency virus gp120 crystal structures, molecular mimicry, and neutralization breadth. Structure. 2005;13(5):755–768. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

