FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development
- PMID: 20876100
- PMCID: PMC2955125
- DOI: 10.1073/pnas.1005366107
FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development
Abstract
Plant RHO GTPases (RAC/ROPs) mediate multiple extracellular signals ranging from hormone to stress and regulate diverse cellular processes important for polarized cell growth, differentiation, development, reproduction, and responses to the environment. They shuttle between the GDP-bound inactive state and the GTP-bound activated state and their activation is predominantly mediated by a family of guanine nucleotide exchange factors (GEFs) referred to as ROPGEFs. Using the Arabidopsis ROPGEF1 as bait, we identified members of a receptor-like kinase (RLK) family as potential upstream regulators for RAC/ROP signaling. NADPH oxidase-derived reactive oxygen species (ROS) are emerging as important regulators for growth and development and play a crucial role in mediating RAC/ROP-regulated root hair development, a polarized cell growth process. We therefore screened T-DNA insertion mutants in these RLKs for root hair defects and found that mutations in one of them, At3g51550 encoding the FERONIA (FER) receptor-like kinase, induced severe root hair defects. We show that the fer phenotypes correlated with reduced levels of active RAC/ROPs and NADPH oxidase-dependent, auxin-regulated ROS accumulation in roots and root hairs and that up-regulating RAC/ROP signaling in fer countered the mutant phenotypes. Taken together, these observations strongly support FER as an upstream regulator for the RAC/ROP-signaled pathway that controls ROS-mediated root hair development. Moreover, FER was pulled down by ROP2 GTPase in a guanine nucleotide-regulated manner implying a dynamic signaling complex involving FER, a ROPGEF, and a RAC/ROP.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
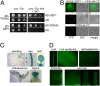
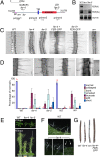
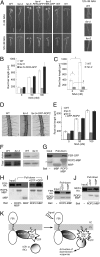
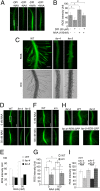
Comment in
-
FERONIA as an upstream receptor kinase for polar cell growth in plants.Proc Natl Acad Sci U S A. 2010 Oct 12;107(41):17461-2. doi: 10.1073/pnas.1013090107. Epub 2010 Oct 6. Proc Natl Acad Sci U S A. 2010. PMID: 20926748 Free PMC article. No abstract available.
Similar articles
-
Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth.New Phytol. 2013 Dec;200(4):1089-101. doi: 10.1111/nph.12432. Epub 2013 Aug 5. New Phytol. 2013. PMID: 23915272
-
Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth.Plant Cell. 2006 Feb;18(2):366-81. doi: 10.1105/tpc.105.036434. Epub 2006 Jan 13. Plant Cell. 2006. PMID: 16415208 Free PMC article.
-
Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in Arabidopsis.Elife. 2015 Jun 8;4:e06587. doi: 10.7554/eLife.06587. Elife. 2015. PMID: 26052747 Free PMC article.
-
FERONIA Receptor Kinase at the Crossroads of Hormone Signaling and Stress Responses.Plant Cell Physiol. 2017 Jul 1;58(7):1143-1150. doi: 10.1093/pcp/pcx048. Plant Cell Physiol. 2017. PMID: 28444222 Review.
-
THESEUS 1, FERONIA and relatives: a family of cell wall-sensing receptor kinases?Curr Opin Plant Biol. 2011 Dec;14(6):632-41. doi: 10.1016/j.pbi.2011.09.001. Epub 2011 Sep 29. Curr Opin Plant Biol. 2011. PMID: 21963060 Review.
Cited by
-
Spatial control of plasma membrane domains: ROP GTPase-based symmetry breaking.Curr Opin Plant Biol. 2012 Dec;15(6):601-7. doi: 10.1016/j.pbi.2012.10.004. Epub 2012 Nov 20. Curr Opin Plant Biol. 2012. PMID: 23177207 Free PMC article. Review.
-
GLABRA2 Regulates Actin Bundling Protein VILLIN1 in Root Hair Growth in Response to Osmotic Stress.Plant Physiol. 2020 Sep;184(1):176-193. doi: 10.1104/pp.20.00480. Epub 2020 Jul 7. Plant Physiol. 2020. PMID: 32636342 Free PMC article.
-
Gene Expression Patterns for Proteins With Lectin Domains in Flax Stem Tissues Are Related to Deposition of Distinct Cell Wall Types.Front Plant Sci. 2021 Apr 26;12:634594. doi: 10.3389/fpls.2021.634594. eCollection 2021. Front Plant Sci. 2021. PMID: 33995436 Free PMC article.
-
Size control in plants--lessons from leaves and flowers.Cold Spring Harb Perspect Biol. 2015 Aug 3;7(8):a019190. doi: 10.1101/cshperspect.a019190. Cold Spring Harb Perspect Biol. 2015. PMID: 26238357 Free PMC article. Review.
-
Hitting the Wall-Sensing and Signaling Pathways Involved in Plant Cell Wall Remodeling in Response to Abiotic Stress.Plants (Basel). 2018 Oct 23;7(4):89. doi: 10.3390/plants7040089. Plants (Basel). 2018. PMID: 30360552 Free PMC article. Review.
References
-
- Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. - PubMed
-
- Nibau C, Wu H-M, Cheung AY. RAC/ROP GTPases: ‘Hubs’ for signal integration and diversification in plants. Trends Plant Sci. 2006;11:309–315. - PubMed
-
- Kost B. Spatial control of Rho (Rac-Rop) signaling in tip-growing plant cells. Trends Cell Biol. 2008;18:119–127. - PubMed
-
- Lavy M, et al. A Novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol. 2007;17:947–952. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

