Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome
- PMID: 20870966
- PMCID: PMC2955092
- DOI: 10.1073/pnas.1012891107
Global effects of the small RNA biogenesis machinery on the Arabidopsis thaliana transcriptome
Abstract
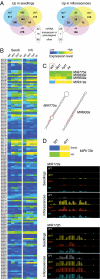
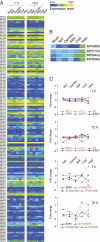
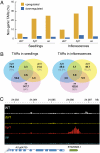
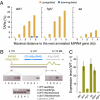
In Arabidopsis thaliana, four different dicer-like (DCL) proteins have distinct but partially overlapping functions in the biogenesis of microRNAs (miRNAs) and siRNAs from longer, noncoding precursor RNAs. To analyze the impact of different components of the small RNA biogenesis machinery on the transcriptome, we subjected dcl and other mutants impaired in small RNA biogenesis to whole-genome tiling array analysis. We compared both protein-coding genes and noncoding transcripts, including most pri-miRNAs, in two tissues and several stress conditions. Our analysis revealed a surprising number of common targets in dcl1 and dcl2 dcl3 dcl4 triple mutants. Furthermore, our results suggest that the DCL1 is not only involved in miRNA action but also contributes to silencing of a subset of transposons, apparently through an effect on DNA methylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Recovery of dicer-like 1-late flowering phenotype by miR172 expressed by the noncanonical DCL4-dependent biogenesis pathway.RNA. 2014 Aug;20(8):1320-7. doi: 10.1261/rna.044966.114. Epub 2014 Jun 25. RNA. 2014. PMID: 24966167 Free PMC article.
-
DAWDLE Interacts with DICER-LIKE Proteins to Mediate Small RNA Biogenesis.Plant Physiol. 2018 Jul;177(3):1142-1151. doi: 10.1104/pp.18.00354. Epub 2018 May 21. Plant Physiol. 2018. PMID: 29784765 Free PMC article.
-
Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways.Plant Cell. 2005 Nov;17(11):2873-85. doi: 10.1105/tpc.105.036608. Epub 2005 Oct 7. Plant Cell. 2005. PMID: 16214897 Free PMC article.
-
Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis.J Plant Res. 2017 Jan;130(1):33-44. doi: 10.1007/s10265-016-0877-1. Epub 2016 Nov 24. J Plant Res. 2017. PMID: 27885504 Review.
-
MicroRNA biogenesis and function in plants.FEBS Lett. 2005 Oct 31;579(26):5923-31. doi: 10.1016/j.febslet.2005.07.071. Epub 2005 Aug 9. FEBS Lett. 2005. PMID: 16144699 Free PMC article. Review.
Cited by
-
Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function.Genome Biol. 2012;13(3):R20. doi: 10.1186/gb-2012-13-3-r20. Genome Biol. 2012. PMID: 22439910 Free PMC article.
-
MicroRNAs regulate the timing of embryo maturation in Arabidopsis.Plant Physiol. 2011 Apr;155(4):1871-84. doi: 10.1104/pp.110.171355. Epub 2011 Feb 17. Plant Physiol. 2011. PMID: 21330492 Free PMC article.
-
microRNA biogenesis, degradation and activity in plants.Cell Mol Life Sci. 2015 Jan;72(1):87-99. doi: 10.1007/s00018-014-1728-7. Epub 2014 Sep 11. Cell Mol Life Sci. 2015. PMID: 25209320 Free PMC article. Review.
-
Abscisic Acid Induces Resistance against Bamboo Mosaic Virus through Argonaute2 and 3.Plant Physiol. 2017 May;174(1):339-355. doi: 10.1104/pp.16.00015. Epub 2017 Mar 7. Plant Physiol. 2017. PMID: 28270624 Free PMC article.
-
Combined Activity of DCL2 and DCL3 Is Crucial in the Defense against Potato Spindle Tuber Viroid.PLoS Pathog. 2016 Oct 12;12(10):e1005936. doi: 10.1371/journal.ppat.1005936. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27732664 Free PMC article.
References
-
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. - PubMed
-
- Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

