Identification of context-dependent motifs by contrasting ChIP binding data
- PMID: 20870645
- PMCID: PMC2971577
- DOI: 10.1093/bioinformatics/btq546
Identification of context-dependent motifs by contrasting ChIP binding data
Abstract
Motivation: DNA binding proteins play crucial roles in the regulation of gene expression. Transcription factors (TFs) activate or repress genes directly while other proteins influence chromatin structure for transcription. Binding sites of a TF exhibit a similar sequence pattern called a motif. However, a one-to-one map does not exist between each TF and motif. Many TFs in a protein family may recognize the same motif with subtle nucleotide differences leading to different binding affinities. Additionally, a particular TF may bind different motifs under certain conditions, for example in the presence of different co-regulators. The availability of genome-wide binding data of multiple collaborative TFs makes it possible to detect such context-dependent motifs.
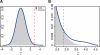
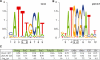
Results: We developed a contrast motif finder (CMF) for the de novo identification of motifs that are differentially enriched in two sets of sequences. Applying this method to a number of TF binding datasets from mouse embryonic stem cells, we demonstrate that CMF achieves substantially higher accuracy than several well-known motif finding methods. By contrasting sequences bound by distinct sets of TFs, CMF identified two different motifs that may be recognized by Oct4 dependent on the presence of another co-regulator and detected subtle motif signals that may be associated with potential competitive binding between Sox2 and Tcf3.
Availability: The software CMF is freely available for academic use at www.stat.ucla.edu/∼zhou/CMF.
Figures
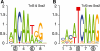
Similar articles
-
Improved linking of motifs to their TFs using domain information.Bioinformatics. 2020 Mar 1;36(6):1655-1662. doi: 10.1093/bioinformatics/btz855. Bioinformatics. 2020. PMID: 31742324 Free PMC article.
-
Co-regulation in embryonic stem cells via context-dependent binding of transcription factors.Bioinformatics. 2013 Sep 1;29(17):2162-8. doi: 10.1093/bioinformatics/btt365. Epub 2013 Jun 21. Bioinformatics. 2013. PMID: 23793746
-
COPS: detecting co-occurrence and spatial arrangement of transcription factor binding motifs in genome-wide datasets.PLoS One. 2012;7(12):e52055. doi: 10.1371/journal.pone.0052055. Epub 2012 Dec 18. PLoS One. 2012. PMID: 23272209 Free PMC article.
-
An algorithmic perspective of de novo cis-regulatory motif finding based on ChIP-seq data.Brief Bioinform. 2018 Sep 28;19(5):1069-1081. doi: 10.1093/bib/bbx026. Brief Bioinform. 2018. PMID: 28334268 Review.
-
A survey of DNA motif finding algorithms.BMC Bioinformatics. 2007 Nov 1;8 Suppl 7(Suppl 7):S21. doi: 10.1186/1471-2105-8-S7-S21. BMC Bioinformatics. 2007. PMID: 18047721 Free PMC article. Review.
Cited by
-
SeAMotE: a method for high-throughput motif discovery in nucleic acid sequences.BMC Genomics. 2014 Oct 23;15(1):925. doi: 10.1186/1471-2164-15-925. BMC Genomics. 2014. PMID: 25341390 Free PMC article.
-
Composing a Tumor Specific Bacterial Promoter.PLoS One. 2016 May 12;11(5):e0155338. doi: 10.1371/journal.pone.0155338. eCollection 2016. PLoS One. 2016. PMID: 27171245 Free PMC article.
-
DREME: motif discovery in transcription factor ChIP-seq data.Bioinformatics. 2011 Jun 15;27(12):1653-9. doi: 10.1093/bioinformatics/btr261. Epub 2011 May 4. Bioinformatics. 2011. PMID: 21543442 Free PMC article.
-
Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program.Development. 2014 Dec;141(23):4537-47. doi: 10.1242/dev.107227. Epub 2014 Oct 30. Development. 2014. PMID: 25359723 Free PMC article.
-
WSMD: weakly-supervised motif discovery in transcription factor ChIP-seq data.Sci Rep. 2017 Jun 12;7(1):3217. doi: 10.1038/s41598-017-03554-7. Sci Rep. 2017. PMID: 28607381 Free PMC article.
References
-
- Barash Y, et al. A simple hyper-geometric approach for discovering putative transcription factor binding sites. Proc. WABI. 2001;1:278–293.
-
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. - PubMed
-
- Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. - PubMed
-
- Chen G, Zhou Q. Heterogeneity in DNA multiple alignments: modeling, inference, and applications in motif finding. Biometrics. 2010;66:694–704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

