Origin of the human malaria parasite Plasmodium falciparum in gorillas
- PMID: 20864995
- PMCID: PMC2997044
- DOI: 10.1038/nature09442
Origin of the human malaria parasite Plasmodium falciparum in gorillas
Abstract
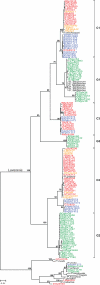
Plasmodium falciparum is the most prevalent and lethal of the malaria parasites infecting humans, yet the origin and evolutionary history of this important pathogen remain controversial. Here we develop a single-genome amplification strategy to identify and characterize Plasmodium spp. DNA sequences in faecal samples from wild-living apes. Among nearly 3,000 specimens collected from field sites throughout central Africa, we found Plasmodium infection in chimpanzees (Pan troglodytes) and western gorillas (Gorilla gorilla), but not in eastern gorillas (Gorilla beringei) or bonobos (Pan paniscus). Ape plasmodial infections were highly prevalent, widely distributed and almost always made up of mixed parasite species. Analysis of more than 1,100 mitochondrial, apicoplast and nuclear gene sequences from chimpanzees and gorillas revealed that 99% grouped within one of six host-specific lineages representing distinct Plasmodium species within the subgenus Laverania. One of these from western gorillas comprised parasites that were nearly identical to P. falciparum. In phylogenetic analyses of full-length mitochondrial sequences, human P. falciparum formed a monophyletic lineage within the gorilla parasite radiation. These findings indicate that P. falciparum is of gorilla origin and not of chimpanzee, bonobo or ancient human origin.
Figures

Comment in
-
Malaria: The gorilla connection.Nature. 2010 Sep 23;467(7314):404-5. doi: 10.1038/467404a. Nature. 2010. PMID: 20864986 No abstract available.
Similar articles
-
Multigenomic Delineation of Plasmodium Species of the Laverania Subgenus Infecting Wild-Living Chimpanzees and Gorillas.Genome Biol Evol. 2016 Jul 2;8(6):1929-39. doi: 10.1093/gbe/evw128. Genome Biol Evol. 2016. PMID: 27289102 Free PMC article.
-
Parasitology. Origin of most deadly human malaria comes out of the mist.Science. 2010 Sep 24;329(5999):1586-7. doi: 10.1126/science.329.5999.1586. Science. 2010. PMID: 20929818 No abstract available.
-
Ecology of malaria infections in western lowland gorillas inhabiting Dzanga Sangha Protected Areas, Central African Republic.Parasitology. 2015 Jun;142(7):890-900. doi: 10.1017/S0031182015000086. Epub 2015 Mar 4. Parasitology. 2015. PMID: 25736484
-
Ape Origins of Human Malaria.Annu Rev Microbiol. 2020 Sep 8;74:39-63. doi: 10.1146/annurev-micro-020518-115628. Annu Rev Microbiol. 2020. PMID: 32905751 Free PMC article. Review.
-
[From malaria parasite point of view--Plasmodium falciparum evolution].Postepy Hig Med Dosw (Online). 2015 Dec 31;69:1519-29. Postepy Hig Med Dosw (Online). 2015. PMID: 27259224 Review. Polish.
Cited by
-
Population Genomic Structure and Recent Evolution of Plasmodium knowlesi, Peninsular Malaysia.Emerg Infect Dis. 2020 Aug;26(8):1749-1758. doi: 10.3201/eid2608.190864. Emerg Infect Dis. 2020. PMID: 32687018 Free PMC article.
-
Resistance to malaria in humans: the impact of strong, recent selection.Malar J. 2012 Oct 22;11:349. doi: 10.1186/1475-2875-11-349. Malar J. 2012. PMID: 23088866 Free PMC article. Review.
-
Use of RNAlater as a preservation method for parasitic coprology studies in wild-living chimpanzees.Exp Parasitol. 2013 Oct;135(2):257-61. doi: 10.1016/j.exppara.2013.07.002. Epub 2013 Jul 11. Exp Parasitol. 2013. PMID: 23850999 Free PMC article.
-
The Genome of Haemoproteus tartakovskyi and Its Relationship to Human Malaria Parasites.Genome Biol Evol. 2016 May 12;8(5):1361-73. doi: 10.1093/gbe/evw081. Genome Biol Evol. 2016. PMID: 27190205 Free PMC article.
-
Role of Pfs47 in the dispersal of ancestral Plasmodium falciparum malaria through adaptation to different anopheline vectors.Proc Natl Acad Sci U S A. 2023 Jan 31;120(5):e2213626120. doi: 10.1073/pnas.2213626120. Epub 2023 Jan 23. Proc Natl Acad Sci U S A. 2023. PMID: 36689648 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R03 AI074778-02/AI/NIAID NIH HHS/United States
- U19 AI 067854/AI/NIAID NIH HHS/United States
- R37 AI050529/AI/NIAID NIH HHS/United States
- T32 AI007245/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 AI 7767/AI/NIAID NIH HHS/United States
- R01 AI058715-06A1/AI/NIAID NIH HHS/United States
- U19 AI067854/AI/NIAID NIH HHS/United States
- R01 AI050529/AI/NIAID NIH HHS/United States
- R01 AI058715/AI/NIAID NIH HHS/United States
- T32 GM008111-13/GM/NIGMS NIH HHS/United States
- T32 GM008111/GM/NIGMS NIH HHS/United States
- R37 AI050529-08/AI/NIAID NIH HHS/United States
- R03 AI074778/AI/NIAID NIH HHS/United States
- R01 AI50529/AI/NIAID NIH HHS/United States
- P30 AI027767-21A1/AI/NIAID NIH HHS/United States
- P30 AI027767/AI/NIAID NIH HHS/United States
- U19 AI067854-06/AI/NIAID NIH HHS/United States
- R01 AI058715-07/AI/NIAID NIH HHS/United States
- T32 AI007245-26/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 I58715/PHS HHS/United States
- R37 AI050529-07/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

