Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC
- PMID: 20863545
- PMCID: PMC2993081
- DOI: 10.1016/j.virol.2010.08.028
Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC
Abstract
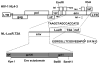
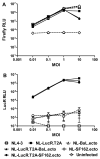
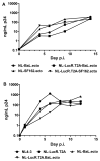
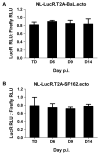
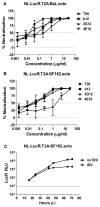
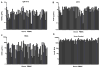
Effective vaccine development for human immunodeficiency virus type 1 (HIV-1) will require assays that ascertain the capacity of vaccine immunogens to elicit neutralizing antibodies (NAb) to diverse HIV-1 strains. To facilitate NAb assessment in peripheral blood mononuclear cell (PBMC)-based assays, we developed an assay-adaptable platform based on a Renilla luciferase (LucR) expressing HIV-1 proviral backbone. LucR was inserted into pNL4-3 DNA, preserving all viral open reading frames. The proviral genome was engineered to facilitate expression of diverse HIV-1 env sequences, allowing analysis in an isogenic background. The resulting Env-IMC-LucR viruses are infectious, and LucR is stably expressed over multiple replications in PBMC. HIV-1 neutralization, targeting TZM-bl cells, was highly correlative comparing virus (LucR) and cell (firefly luciferase) readouts. In PBMC, NAb activity can be analyzed either within a single or multiple cycles of replication. These results represent advancement toward a standardizable PBMC-based neutralization assay for assessing HIV-1 vaccine immunogen efficacy.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures


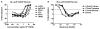
Similar articles
-
Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies.J Virol. 2014 Mar;88(5):2489-507. doi: 10.1128/JVI.02853-13. Epub 2013 Dec 18. J Virol. 2014. PMID: 24352443 Free PMC article.
-
Development of a luciferase based viral inhibition assay to evaluate vaccine induced CD8 T-cell responses.J Immunol Methods. 2014 Jul;409:161-73. doi: 10.1016/j.jim.2013.11.021. Epub 2013 Nov 28. J Immunol Methods. 2014. PMID: 24291126 Free PMC article.
-
Impact of HIV-1 backbone on neutralization sensitivity: neutralization profiles of heterologous envelope glycoproteins expressed in native subtype C and CRF01_AE backbone.PLoS One. 2013 Nov 29;8(11):e76104. doi: 10.1371/journal.pone.0076104. eCollection 2013. PLoS One. 2013. PMID: 24312165 Free PMC article.
-
Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination.Virology. 2008 Jun 5;375(2):315-20. doi: 10.1016/j.virol.2008.02.007. Epub 2008 Mar 25. Virology. 2008. PMID: 18367229 Review.
-
New virologic reagents for neutralizing antibody assays.Curr Opin HIV AIDS. 2009 Sep;4(5):418-25. doi: 10.1097/COH.0b013e32832f011e. Curr Opin HIV AIDS. 2009. PMID: 20048706 Review.
Cited by
-
Vaccine-induced plasma IgA specific for the C1 region of the HIV-1 envelope blocks binding and effector function of IgG.Proc Natl Acad Sci U S A. 2013 May 28;110(22):9019-24. doi: 10.1073/pnas.1301456110. Epub 2013 May 9. Proc Natl Acad Sci U S A. 2013. PMID: 23661056 Free PMC article.
-
Magnitude and breadth of the neutralizing antibody response in the RV144 and Vax003 HIV-1 vaccine efficacy trials.J Infect Dis. 2012 Aug 1;206(3):431-41. doi: 10.1093/infdis/jis367. Epub 2012 May 25. J Infect Dis. 2012. PMID: 22634875 Free PMC article. Clinical Trial.
-
In vivo killing of primary HIV-infected cells by peripheral-injected early memory-enriched anti-HIV duoCAR T cells.JCI Insight. 2022 Nov 8;7(21):e161698. doi: 10.1172/jci.insight.161698. JCI Insight. 2022. PMID: 36345941 Free PMC article.
-
Potent functional antibody responses elicited by HIV-I DNA priming and boosting with heterologous HIV-1 recombinant MVA in healthy Tanzanian adults.PLoS One. 2015 Apr 14;10(4):e0118486. doi: 10.1371/journal.pone.0118486. eCollection 2015. PLoS One. 2015. PMID: 25874723 Free PMC article. Clinical Trial.
-
The Vaginal Acquisition and Dissemination of HIV-1 Infection in a Novel Transgenic Mouse Model Is Facilitated by Coinfection with Herpes Simplex Virus 2 and Is Inhibited by Microbicide Treatment.J Virol. 2015 Sep;89(18):9559-70. doi: 10.1128/JVI.01326-15. Epub 2015 Jul 8. J Virol. 2015. PMID: 26157126 Free PMC article.
References
-
- Baba TW, Liska V, Hofmann-Lehmann R, Vlasak J, Xu W, Ayehunie S, Cavacini LA, Posner MR, Katinger H, Stiegler G, Bernacky BJ, Rizvi TA, Schmidt R, Hill LR, Keeling ME, Lu Y, Wright JE, Chou TC, Ruprecht RM. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat Med. 2000;6(2):200–6. - PubMed
-
- Bentham M, Mazaleyrat S, Harris M. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J Gen Virol. 2006;87(Pt 3):563–71. - PubMed
-
- Binley JM, Lybarger EA, Crooks ET, Seaman MS, Gray E, Davis KL, Decker JM, Wycuff D, Harris L, Hawkins N, Wood B, Nathe C, Richman D, Tomaras GD, Bibollet-Ruche F, Robinson JE, Morris L, Shaw GM, Montefiori DC, Mascola JR. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol. 2008;82(23):11651–68. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

