The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I
- PMID: 20855291
- PMCID: PMC3035449
- DOI: 10.1093/nar/gkq822
The geometry of DNA supercoils modulates the DNA cleavage activity of human topoisomerase I
Abstract
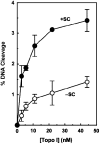
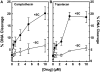
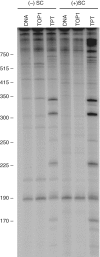
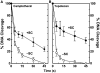
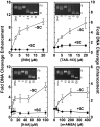
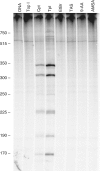
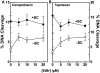
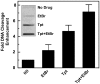
Human topoisomerase I plays an important role in removing positive DNA supercoils that accumulate ahead of replication forks. It also is the target for camptothecin-based anticancer drugs that act by increasing levels of topoisomerase I-mediated DNA scission. Evidence suggests that cleavage events most likely to generate permanent genomic damage are those that occur ahead of DNA tracking systems. Therefore, it is important to characterize the ability of topoisomerase I to cleave positively supercoiled DNA. Results confirm that the human enzyme maintains higher levels of cleavage with positively as opposed to negatively supercoiled substrates in the absence or presence of anticancer drugs. Enhanced drug efficacy on positively supercoiled DNA is due primarily to an increase in baseline levels of cleavage. Sites of topoisomerase I-mediated DNA cleavage do not appear to be affected by supercoil geometry. However, rates of ligation are slower with positively supercoiled substrates. Finally, intercalators enhance topoisomerase I-mediated cleavage of negatively supercoiled substrates but not positively supercoiled or linear DNA. We suggest that these compounds act by altering the perceived topological state of the double helix, making underwound DNA appear to be overwound to the enzyme, and propose that these compounds be referred to as 'topological poisons of topoisomerase I'.
Figures
Similar articles
-
The geometry of DNA supercoils modulates topoisomerase-mediated DNA cleavage and enzyme response to anticancer drugs.Biochemistry. 2006 Mar 7;45(9):3040-50. doi: 10.1021/bi051987q. Biochemistry. 2006. PMID: 16503659 Free PMC article.
-
Recognition of DNA Supercoil Geometry by Mycobacterium tuberculosis Gyrase.Biochemistry. 2017 Oct 10;56(40):5440-5448. doi: 10.1021/acs.biochem.7b00681. Epub 2017 Sep 25. Biochemistry. 2017. PMID: 28921956 Free PMC article.
-
Activities of gyrase and topoisomerase IV on positively supercoiled DNA.Nucleic Acids Res. 2017 Sep 19;45(16):9611-9624. doi: 10.1093/nar/gkx649. Nucleic Acids Res. 2017. PMID: 28934496 Free PMC article.
-
Oligonucleotide-Recognizing Topoisomerase Inhibitors (OTIs): Precision Gene Editors for Neurodegenerative Diseases?Int J Mol Sci. 2022 Sep 29;23(19):11541. doi: 10.3390/ijms231911541. Int J Mol Sci. 2022. PMID: 36232843 Free PMC article. Review.
-
Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs.Curr Med Chem. 2010;17(35):4270-90. doi: 10.2174/092986710793361252. Curr Med Chem. 2010. PMID: 20939813 Review.
Cited by
-
Unraveling topoisomerase IA gate dynamics in presence of PPEF and its preclinical evaluation against multidrug-resistant pathogens.Commun Biol. 2023 Feb 18;6(1):195. doi: 10.1038/s42003-023-04412-1. Commun Biol. 2023. PMID: 36807602 Free PMC article.
-
A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity.Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):16125-30. doi: 10.1073/pnas.1206480109. Epub 2012 Sep 18. Proc Natl Acad Sci U S A. 2012. PMID: 22991469 Free PMC article.
-
Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription.Mutat Res. 2013 Mar-Apr;743-744:89-96. doi: 10.1016/j.mrfmmm.2012.12.002. Epub 2012 Dec 19. Mutat Res. 2013. PMID: 23261817 Free PMC article.
-
Neutral Porphyrin Derivative Exerts Anticancer Activity by Targeting Cellular Topoisomerase I (Top1) and Promotes Apoptotic Cell Death without Stabilizing Top1-DNA Cleavage Complexes.J Med Chem. 2018 Feb 8;61(3):804-817. doi: 10.1021/acs.jmedchem.7b01297. Epub 2018 Jan 11. J Med Chem. 2018. PMID: 29290109 Free PMC article.
-
The dynamic interplay between DNA topoisomerases and DNA topology.Biophys Rev. 2016 Nov;8(Suppl 1):101-111. doi: 10.1007/s12551-016-0240-8. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28510219 Free PMC article. Review.
References
-
- Wang JC. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. - PubMed
-
- Cozzarelli NR, Wang JC, editors. DNA Topology and its Biological Effects. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 1990.
-
- Kanaar R, Cozzarelli NR. Roles of supercoiled DNA structure in DNA transactions. Curr. Opin. Struct. Biol. 1992;2:369–379.
-
- Bates AD, Maxwell A. DNA Topology. 2nd edn. Oxford: Oxford University Press; 2005.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

