Conserved mutation of Epstein-Barr virus-encoded BamHI-A Rightward Frame-1 (BARF1) gene in Indonesian nasopharyngeal carcinoma
- PMID: 20849661
- PMCID: PMC2949665
- DOI: 10.1186/1750-9378-5-16
Conserved mutation of Epstein-Barr virus-encoded BamHI-A Rightward Frame-1 (BARF1) gene in Indonesian nasopharyngeal carcinoma
Abstract
Background: BamHI-A rightward frame-1 (BARF1) is a carcinoma-specific Epstein-Barr virus (EBV) encoded oncogene. Here we describe the BARF1 sequence diversity in nasopharyngeal carcinoma (NPC), other EBV-related diseases and Indonesian healthy EBV carriers in relation to EBV genotype, viral load and serology markers. Nasopharyngeal brushings from 56 NPC cases, blood or tissue from 15 other EBV-related disorders, spontaneous B cell lines (LCL) from 5 Indonesian healthy individuals and several prototype EBV isolates were analysed by PCR-direct sequencing.
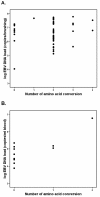
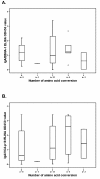
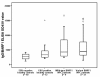
Results: Most NPC isolates revealed specific BARF1 nucleotide changes compared to prototype B95-8 virus. At the protein level these mutations resulted in 3 main substitutions (V29A, W72G, H130R), which are not considered to cause gross tertiary structure alterations in the hexameric BARF1 protein. At least one amino acid conversion was detected in 80.3% of NPC samples compared to 33.3% of non-NPC samples (p < 0.001) and 40.0% of healthy LCLs (p = 0.074). NPC isolates also showed more frequent codon mutation than non-NPC samples. EBV strain typing revealed most isolates as EBV type 1. The viral load of either NPC or non-NPC samples was high, but only in non- NPC group it related to a particular BARF1 variant. Serology on NPC sera using IgA/EBNA-1 ELISA, IgA/VCA-p18 ELISA and immunoblot score showed no relation with BARF1 sequence diversity (p = 0.802, 0.382 and 0.058, respectively). NPC patients had variable antibody reactivity against purified hexameric NPC-derived BARF1 irrespective of the endogenous BARF1 sequence.
Conclusion: The sequence variation of BARF1 observed in Indonesian NPC patients and controls may reflect a natural selection of EBV strains unlikely to be predisposing to carcinogenesis. The conserved nature of BARF1 may reflect an important role in EBV (epithelial) persistence.
Figures

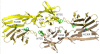
Similar articles
-
Role of BamHI-A Rightward Frame 1 in Epstein-Barr Virus-Associated Epithelial Malignancies.Biology (Basel). 2020 Dec 11;9(12):461. doi: 10.3390/biology9120461. Biology (Basel). 2020. PMID: 33322292 Free PMC article. Review.
-
Unique variations of Epstein-Barr virus-encoded BARF1 gene in nasopharyngeal carcinoma biopsies.Virus Res. 2012 Jun;166(1-2):23-30. doi: 10.1016/j.virusres.2012.02.022. Epub 2012 Mar 1. Virus Res. 2012. PMID: 22406129
-
Purified hexameric Epstein-Barr virus-encoded BARF1 protein for measuring anti-BARF1 antibody responses in nasopharyngeal carcinoma patients.Clin Vaccine Immunol. 2011 Feb;18(2):298-304. doi: 10.1128/CVI.00193-10. Epub 2010 Dec 1. Clin Vaccine Immunol. 2011. PMID: 21123521 Free PMC article.
-
Sequence variations of Epstein-Barr virus-encoded BARF1 gene in nasopharyngeal carcinomas and healthy donors from southern and northern China.J Med Virol. 2018 Oct;90(10):1629-1635. doi: 10.1002/jmv.25233. Epub 2018 Jun 13. J Med Virol. 2018. PMID: 29797589
-
The Therapeutic Potential of Targeting BARF1 in EBV-Associated Malignancies.Cancers (Basel). 2020 Jul 17;12(7):1940. doi: 10.3390/cancers12071940. Cancers (Basel). 2020. PMID: 32708965 Free PMC article. Review.
Cited by
-
Nasopharyngeal Carcinoma Progression: Accumulating Genomic Instability and Persistent Epstein-Barr Virus Infection.Curr Oncol. 2022 Aug 23;29(9):6035-6052. doi: 10.3390/curroncol29090475. Curr Oncol. 2022. PMID: 36135044 Free PMC article. Review.
-
Genomic assays for Epstein-Barr virus-positive gastric adenocarcinoma.Exp Mol Med. 2015 Jan 23;47(1):e134. doi: 10.1038/emm.2014.93. Exp Mol Med. 2015. PMID: 25613731 Free PMC article. Review.
-
Natural Variations in BRLF1 Promoter Contribute to the Elevated Reactivation Level of Epstein-Barr Virus in Endemic Areas of Nasopharyngeal Carcinoma.EBioMedicine. 2018 Nov;37:101-109. doi: 10.1016/j.ebiom.2018.10.065. Epub 2018 Nov 9. EBioMedicine. 2018. PMID: 30420297 Free PMC article. Clinical Trial.
-
Comprehensive insight into altered host cell-signaling cascades upon Helicobacter pylori and Epstein-Barr virus infections in cancer.Arch Microbiol. 2023 Jun 13;205(7):262. doi: 10.1007/s00203-023-03598-6. Arch Microbiol. 2023. PMID: 37310490 Review.
-
Role of BamHI-A Rightward Frame 1 in Epstein-Barr Virus-Associated Epithelial Malignancies.Biology (Basel). 2020 Dec 11;9(12):461. doi: 10.3390/biology9120461. Biology (Basel). 2020. PMID: 33322292 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

