Improving the accuracy of predicting secondary structure for aligned RNA sequences
- PMID: 20843778
- PMCID: PMC3025558
- DOI: 10.1093/nar/gkq792
Improving the accuracy of predicting secondary structure for aligned RNA sequences
Abstract
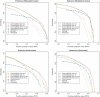
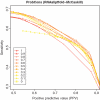
Considerable attention has been focused on predicting the secondary structure for aligned RNA sequences since it is useful not only for improving the limiting accuracy of conventional secondary structure prediction but also for finding non-coding RNAs in genomic sequences. Although there exist many algorithms of predicting secondary structure for aligned RNA sequences, further improvement of the accuracy is still awaited. In this article, toward improving the accuracy, a theoretical classification of state-of-the-art algorithms of predicting secondary structure for aligned RNA sequences is presented. The classification is based on the viewpoint of maximum expected accuracy (MEA), which has been successfully applied in various problems in bioinformatics. The classification reveals several disadvantages of the current algorithms but we propose an improvement of a previously introduced algorithm (CentroidAlifold). Finally, computational experiments strongly support the theoretical classification and indicate that the improved CentroidAlifold substantially outperforms other algorithms.
Figures


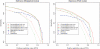
Similar articles
-
RNA secondary structure prediction from multi-aligned sequences.Methods Mol Biol. 2015;1269:17-38. doi: 10.1007/978-1-4939-2291-8_2. Methods Mol Biol. 2015. PMID: 25577370
-
RNA structural alignments, part II: non-Sankoff approaches for structural alignments.Methods Mol Biol. 2014;1097:291-301. doi: 10.1007/978-1-62703-709-9_14. Methods Mol Biol. 2014. PMID: 24639165
-
CentroidAlign: fast and accurate aligner for structured RNAs by maximizing expected sum-of-pairs score.Bioinformatics. 2009 Dec 15;25(24):3236-43. doi: 10.1093/bioinformatics/btp580. Epub 2009 Oct 6. Bioinformatics. 2009. PMID: 19808876
-
Energy-based RNA consensus secondary structure prediction in multiple sequence alignments.Methods Mol Biol. 2014;1097:125-41. doi: 10.1007/978-1-62703-709-9_7. Methods Mol Biol. 2014. PMID: 24639158 Review.
-
Computational analysis of RNA structures with chemical probing data.Methods. 2015 Jun;79-80:60-6. doi: 10.1016/j.ymeth.2015.02.003. Epub 2015 Feb 14. Methods. 2015. PMID: 25687190 Free PMC article. Review.
Cited by
-
CompaRNA: a server for continuous benchmarking of automated methods for RNA secondary structure prediction.Nucleic Acids Res. 2013 Apr;41(7):4307-23. doi: 10.1093/nar/gkt101. Epub 2013 Feb 21. Nucleic Acids Res. 2013. PMID: 23435231 Free PMC article.
-
aliFreeFold: an alignment-free approach to predict secondary structure from homologous RNA sequences.Bioinformatics. 2018 Jul 1;34(13):i70-i78. doi: 10.1093/bioinformatics/bty234. Bioinformatics. 2018. PMID: 29949960 Free PMC article.
-
Multiple sequence alignment-based RNA language model and its application to structural inference.Nucleic Acids Res. 2024 Jan 11;52(1):e3. doi: 10.1093/nar/gkad1031. Nucleic Acids Res. 2024. PMID: 37941140 Free PMC article.
-
A classification of bioinformatics algorithms from the viewpoint of maximizing expected accuracy (MEA).J Comput Biol. 2012 May;19(5):532-49. doi: 10.1089/cmb.2011.0197. Epub 2012 Feb 7. J Comput Biol. 2012. PMID: 22313125 Free PMC article. Review.
-
LinAliFold and CentroidLinAliFold: fast RNA consensus secondary structure prediction for aligned sequences using beam search methods.Bioinform Adv. 2022 Oct 22;2(1):vbac078. doi: 10.1093/bioadv/vbac078. eCollection 2022. Bioinform Adv. 2022. PMID: 36699418 Free PMC article.
References
-
- Bernhart SH, Hofacker IL. From consensus structure prediction to RNA gene finding. Brief. Funct. Genomic Proteomic. 2009;8:461–471. - PubMed
-
- Hofacker I, Fontana W, Stadler P, Bonhoeffer S, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh. Chem. 1994;125:167–188.

