Prion strain interactions are highly selective
- PMID: 20826672
- PMCID: PMC2951977
- DOI: 10.1523/JNEUROSCI.2417-10.2010
Prion strain interactions are highly selective
Abstract
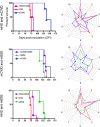
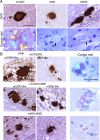
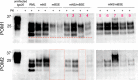
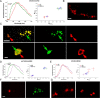
Various misfolded and aggregated neuronal proteins commonly coexist in neurodegenerative disease, but whether the proteins coaggregate and alter the disease pathogenesis is unclear. Here, we used mixtures of distinct prion strains, which are believed to differ in conformation, to test the hypothesis that two different aggregates interact and change the disease in vivo. We tracked two prion strains in mice histopathologically and biochemically, as well as by spectral analysis of plaque-bound PTAA (polythiophene acetic acid), a conformation-sensitive fluorescent amyloid ligand. We found that prion strains interacted in a highly selective and strain-specific manner, with (1) no interaction, (2) hybrid plaque formation, or (3) blockage of one strain by a second (interference). The hybrid plaques were maintained on additional passage in vivo and each strain seemed to maintain its original conformational properties, suggesting that one strain served only as a scaffold for aggregation of the second strain. These findings not only further our understanding of prion strain interactions but also directly demonstrate interactions that may occur in other protein aggregate mixtures.
Figures

Similar articles
-
Multimodal fluorescence microscopy of prion strain specific PrP deposits stained by thiophene-based amyloid ligands.Prion. 2014;8(4):319-29. doi: 10.4161/pri.29239. Epub 2014 Nov 1. Prion. 2014. PMID: 25495506 Free PMC article.
-
Molecular biology and pathology of scrapie and the prion diseases of humans.Brain Pathol. 1991 Jul;1(4):297-310. doi: 10.1111/j.1750-3639.1991.tb00673.x. Brain Pathol. 1991. PMID: 1669719 Review.
-
Overexpression of chimaeric murine/ovine PrP (A136H154Q171) in transgenic mice facilitates transmission and differentiation of ruminant prions.J Gen Virol. 2013 Nov;94(Pt 11):2577-2586. doi: 10.1099/vir.0.051581-0. Epub 2013 Jun 12. J Gen Virol. 2013. PMID: 23761404
-
Prion encephalopathies of animals and humans.Dev Biol Stand. 1993;80:31-44. Dev Biol Stand. 1993. PMID: 8270114 Review.
-
Transgenic models of prion disease.Arch Virol Suppl. 2000;(16):113-24. doi: 10.1007/978-3-7091-6308-5_10. Arch Virol Suppl. 2000. PMID: 11214913 Review.
Cited by
-
Environmental and host factors that contribute to prion strain evolution.Acta Neuropathol. 2021 Jul;142(1):5-16. doi: 10.1007/s00401-021-02310-6. Epub 2021 Apr 25. Acta Neuropathol. 2021. PMID: 33899132 Free PMC article. Review.
-
Cerebral vascular amyloid seeds drive amyloid β-protein fibril assembly with a distinct anti-parallel structure.Nat Commun. 2016 Nov 21;7:13527. doi: 10.1038/ncomms13527. Nat Commun. 2016. PMID: 27869115 Free PMC article.
-
The amyloid state of proteins in human diseases.Cell. 2012 Mar 16;148(6):1188-203. doi: 10.1016/j.cell.2012.02.022. Cell. 2012. PMID: 22424229 Free PMC article. Review.
-
Different misfolding mechanisms converge on common conformational changes: human prion protein pathogenic mutants Y218N and E196K.Prion. 2014 Jan-Feb;8(1):125-35. doi: 10.4161/pri.27807. Prion. 2014. PMID: 24509603 Free PMC article.
-
Luminescent conjugated oligothiophenes for sensitive fluorescent assignment of protein inclusion bodies.Chembiochem. 2013 Mar 18;14(5):607-16. doi: 10.1002/cbic.201200731. Epub 2013 Feb 28. Chembiochem. 2013. PMID: 23450708 Free PMC article.
References
-
- Andersson MR, Berggren M, Olinga T, Hjertberg T, Inganas O, Wennerstrom O. Improved photoluminescence efficiency of films from conjugated polymers. Synth Met. 1997;85:1383–1384.
-
- Bartz JC, Aiken JM, Bessen RA. Delay in onset of prion disease for the HY strain of transmissible mink encephalopathy as a result of prior peripheral inoculation with the replication-deficient DY strain. J Gen Virol. 2004;85:265–273. - PubMed
-
- Berggren M, Bergman P, Fagerstrom J, Inganas O, Andersson M, Weman H, Granstrom M, Stafstrom S, Wennerstrom O, Hjertberg T. Controlling inter-chain and intra-chain excitations of a poly(thiophene) derivative in thin films. Chem Phys Lett. 1999;304:84–90.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
