Medulloblastoma comprises four distinct molecular variants
- PMID: 20823417
- PMCID: PMC4874239
- DOI: 10.1200/JCO.2009.27.4324
Medulloblastoma comprises four distinct molecular variants
Abstract
Purpose: Recent genomic approaches have suggested the existence of multiple distinct subtypes of medulloblastoma. We studied a large cohort of medulloblastomas to determine how many subgroups of the disease exist, how they differ, and the extent of overlap between subgroups.
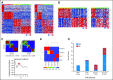
Methods: We determined gene expression profiles and DNA copy number aberrations for 103 primary medulloblastomas. Bioinformatic tools were used for class discovery of medulloblastoma subgroups based on the most informative genes in the data set. Immunohistochemistry for subgroup-specific signature genes was used to determine subgroup affiliation for 294 nonoverlapping medulloblastomas on two independent tissue microarrays.
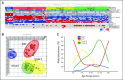
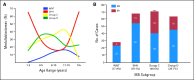
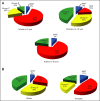
Results: Multiple unsupervised analyses of transcriptional profiles identified the following four distinct, nonoverlapping molecular variants: WNT, SHH, group C, and group D. Supervised analysis of these four subgroups revealed significant subgroup-specific demographics, histology, metastatic status, and DNA copy number aberrations. Immunohistochemistry for DKK1 (WNT), SFRP1 (SHH), NPR3 (group C), and KCNA1 (group D) could reliably and uniquely classify formalin-fixed medulloblastomas in approximately 98% of patients. Group C patients (NPR3-positive tumors) exhibited a significantly diminished progression-free and overall survival irrespective of their metastatic status.
Conclusion: Our integrative genomics approach to a large cohort of medulloblastomas has identified four disparate subgroups with distinct demographics, clinical presentation, transcriptional profiles, genetic abnormalities, and clinical outcome. Medulloblastomas can be reliably assigned to subgroups through immunohistochemistry, thereby making medulloblastoma subclassification widely available. Future research on medulloblastoma and the development of clinical trials should take into consideration these four distinct types of medulloblastoma.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures

Comment in
-
Molecular biology of medulloblastoma: bridging the gap between research and practice.Expert Rev Neurother. 2011 Apr;11(4):491-4. doi: 10.1586/ern.11.22. Expert Rev Neurother. 2011. PMID: 21469921
Similar articles
-
Subgroup-specific alternative splicing in medulloblastoma.Acta Neuropathol. 2012 Apr;123(4):485-499. doi: 10.1007/s00401-012-0959-7. Epub 2012 Feb 23. Acta Neuropathol. 2012. PMID: 22358458 Free PMC article.
-
Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features.PLoS One. 2008 Aug 28;3(8):e3088. doi: 10.1371/journal.pone.0003088. PLoS One. 2008. PMID: 18769486 Free PMC article.
-
A patient with medulloblastoma in its early developmental stage.J Neurosurg Pediatr. 2014 Dec;14(6):615-20. doi: 10.3171/2014.8.PEDS13590. Epub 2014 Oct 10. J Neurosurg Pediatr. 2014. PMID: 25303160
-
Risk stratification of childhood medulloblastoma in the molecular era: the current consensus.Acta Neuropathol. 2016 Jun;131(6):821-31. doi: 10.1007/s00401-016-1569-6. Epub 2016 Apr 4. Acta Neuropathol. 2016. PMID: 27040285 Free PMC article. Review.
-
Medulloblastoma in the age of molecular subgroups: a review.J Neurosurg Pediatr. 2019 Oct 1;24(4):353-363. doi: 10.3171/2019.5.PEDS18381. Epub 2019 Oct 1. J Neurosurg Pediatr. 2019. PMID: 31574483 Review.
Cited by
-
Aptamers: Novel Therapeutics and Potential Role in Neuro-Oncology.Cancers (Basel). 2020 Oct 9;12(10):2889. doi: 10.3390/cancers12102889. Cancers (Basel). 2020. PMID: 33050158 Free PMC article. Review.
-
Sox2: regulation of expression and contribution to brain tumors.CNS Oncol. 2016 Jul;5(3):159-73. doi: 10.2217/cns-2016-0001. Epub 2016 May 27. CNS Oncol. 2016. PMID: 27230973 Free PMC article. Review.
-
DDX3X Suppresses the Susceptibility of Hindbrain Lineages to Medulloblastoma.Dev Cell. 2020 Aug 24;54(4):455-470.e5. doi: 10.1016/j.devcel.2020.05.027. Epub 2020 Jun 17. Dev Cell. 2020. PMID: 32553121 Free PMC article.
-
Functional genomics identifies drivers of medulloblastoma dissemination.Cancer Res. 2012 Oct 1;72(19):4944-53. doi: 10.1158/0008-5472.CAN-12-1629. Epub 2012 Aug 8. Cancer Res. 2012. PMID: 22875024 Free PMC article.
-
Typical Pediatric Brain Tumors Occurring in Adults-Differences in Management and Outcome.Biomedicines. 2021 Mar 30;9(4):356. doi: 10.3390/biomedicines9040356. Biomedicines. 2021. PMID: 33808415 Free PMC article. Review.
References
-
- Ray A, Ho M, Ma J, et al. A clinicobiological model predicting survival in medulloblastoma. Clin Cancer Res. 2004;10:7613–7620. - PubMed
-
- Gilbertson RJ, Perry RH, Kelly PJ, et al. Prognostic significance of HER2 and HER4 coexpression in childhood medulloblastoma. Cancer Res. 1997;57:3272–3280. - PubMed
-
- Grotzer MA, Janss AJ, Fung K, et al. TrkC expression predicts good clinical outcome in primitive neuroectodermal brain tumors. J Clin Oncol. 2000;18:1027–1035. - PubMed
-
- Clifford SC, Lusher ME, Lindsey JC, et al. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5:2666–2670. - PubMed
-
- Ellison DW, Onilude OE, Lindsey JC, et al. Beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: The United Kingdom Children's Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23:7951–7957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

