Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers
- PMID: 20823146
- PMCID: PMC2940983
- DOI: 10.1158/1078-0432.CCR-10-0523
Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers
Abstract
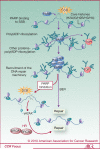
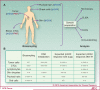
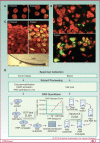
Tumor cells are often deficient in DNA damage response (DDR) pathways, and anticancer therapies are commonly based on genotoxic treatments using radiation and/or drugs that damage DNA directly or interfere with DNA metabolism, leading to the formation of DNA double-strand breaks (DSB), and ultimately to cell death. Because DSBs induce the phosphorylation of histone H2AX (γH2AX) in the chromatin flanking the break site, an antibody directed against γH2AX can be employed to measure DNA damage levels before and after patient treatment. Poly(ADP-ribose) polymerases (PARP1 and PARP2) are also activated by DNA damage, and PARP inhibitors show promising activity in cancers with defective homologous recombination (HR) pathways for DSB repair. Ongoing clinical trials are testing combinations of PARP inhibitors with DNA damaging agents. Poly(ADP-ribosylation), abbreviated as PAR, can be measured in clinical samples and used to determine the efficiency of PARP inhibitors. This review summarizes the roles of γH2AX and PAR in the DDR, and their use as biomarkers to monitor drug response and guide clinical trials, especially phase 0 clinical trials. We also discuss the choices of relevant samples for γH2AX and PAR analyses.
©2010 AACR.
Figures

Similar articles
-
Poly(ADP-ribose) polymerases PARP1 and PARP2 modulate topoisomerase II beta (TOP2B) function during chromatin condensation in mouse spermiogenesis.Biol Reprod. 2011 May;84(5):900-9. doi: 10.1095/biolreprod.110.090035. Epub 2011 Jan 12. Biol Reprod. 2011. PMID: 21228215 Free PMC article.
-
Effect of mild temperature shift on poly(ADP-ribose) and γH2AX levels in cultured cells.Biochem Biophys Res Commun. 2016 Aug 5;476(4):594-599. doi: 10.1016/j.bbrc.2016.06.001. Epub 2016 Jun 2. Biochem Biophys Res Commun. 2016. PMID: 27262441 Free PMC article.
-
Common and unique genetic interactions of the poly(ADP-ribose) polymerases PARP1 and PARP2 with DNA double-strand break repair pathways.DNA Repair (Amst). 2016 Sep;45:56-62. doi: 10.1016/j.dnarep.2016.06.001. Epub 2016 Jun 16. DNA Repair (Amst). 2016. PMID: 27373144 Free PMC article.
-
The role of poly(ADP-ribosyl)ation in DNA damage response and cancer chemotherapy.Oncogene. 2015 Jun;34(26):3349-56. doi: 10.1038/onc.2014.295. Epub 2014 Sep 15. Oncogene. 2015. PMID: 25220415 Free PMC article. Review.
-
Relation between carcinogenesis, chromatin structure and poly(ADP-ribosylation) (review).Anticancer Res. 1991 Mar-Apr;11(2):489-527. Anticancer Res. 1991. PMID: 1905900 Review.
Cited by
-
A Review of Radiation-Induced Alterations of Multi-Omic Profiles, Radiation Injury Biomarkers, and Countermeasures.Radiat Res. 2023 Jan 1;199(1):89-111. doi: 10.1667/RADE-21-00187.1. Radiat Res. 2023. PMID: 36368026 Free PMC article. Review.
-
Drug release patterns and cytotoxicity of PEG-poly(aspartate) block copolymer micelles in cancer cells.Pharm Res. 2012 Jul;29(7):1755-67. doi: 10.1007/s11095-012-0697-5. Epub 2012 Feb 10. Pharm Res. 2012. PMID: 22322898
-
Increased single-strand annealing rather than non-homologous end-joining predicts hereditary ovarian carcinoma.Oncotarget. 2017 Oct 9;8(58):98660-98676. doi: 10.18632/oncotarget.21720. eCollection 2017 Nov 17. Oncotarget. 2017. PMID: 29228718 Free PMC article.
-
Histone deacetylases 1 and 2 cooperate in regulating BRCA1, CHK1, and RAD51 expression in acute myeloid leukemia cells.Oncotarget. 2017 Jan 24;8(4):6319-6329. doi: 10.18632/oncotarget.14062. Oncotarget. 2017. PMID: 28030834 Free PMC article.
-
AKT signaling as a novel factor associated with in vitro resistance of human AML to gemtuzumab ozogamicin.PLoS One. 2013;8(1):e53518. doi: 10.1371/journal.pone.0053518. Epub 2013 Jan 8. PLoS One. 2013. PMID: 23320091 Free PMC article.
References
-
- Kummar S, Kinders R, Rubinstein L, et al. Compressing drug development timelines in oncology using phase ‘0’ trials. Nat Rev Cancer. 2007;7:131–9. - PubMed
-
- Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nat Rev Drug Discov. 2003;2:566–80. - PubMed
-
- Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–9. - PubMed
-
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

