Acetylation of RNA processing proteins and cell cycle proteins in mitosis
- PMID: 20812760
- PMCID: PMC2935306
- DOI: 10.1021/pr100281h
Acetylation of RNA processing proteins and cell cycle proteins in mitosis
Abstract
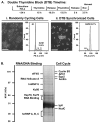
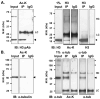
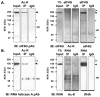
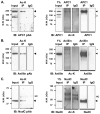
Mitosis is a highly regulated process in which errors can lead to genomic instability, a hallmark of cancer. During this phase of the cell cycle, transcription is silent and RNA translation is inhibited. Thus, mitosis is largely driven by post-translational modification of proteins, including phosphorylation, methylation, ubiquitination, and sumoylation. Here, we show that protein acetylation is prevalent during mitosis. To identify proteins that are acetylated, we synchronized HeLa cells in early prometaphase and immunoprecipitated lysine-acetylated proteins with antiacetyl-lysine antibody. The immunoprecipitated proteins were identified by LC-ESI-MS/MS analysis. These include proteins involved in RNA translation, RNA processing, cell cycle regulation, transcription, chaperone function, DNA damage repair, metabolism, immune response, and cell structure. Immunoprecipitation followed by Western blot analyses confirmed that two RNA processing proteins, eIF4G and RNA helicase A, and several cell cycle proteins, including APC1, anillin, and NudC, were acetylated in mitosis. We further showed that acetylation of APC1 and NudC was enhanced by apicidin treatment, suggesting that their acetylation was regulated by histone deacetylase. Moreover, treating mitotic cells with apicidin or trichostatin A induced spindle abnormalities and cytokinesis failure. These studies suggest that protein acetylation/deacetylation is likely an important regulatory mechanism in mitosis.
Figures

Similar articles
-
Identifying acetylated proteins in mitosis.Methods Mol Biol. 2012;909:181-204. doi: 10.1007/978-1-61779-959-4_13. Methods Mol Biol. 2012. PMID: 22903717
-
NudC deacetylation regulates mitotic progression.PLoS One. 2013 Sep 19;8(9):e73841. doi: 10.1371/journal.pone.0073841. eCollection 2013. PLoS One. 2013. PMID: 24069238 Free PMC article.
-
Inhibition of histone deacetylase activity increases chromosomal instability by the aberrant regulation of mitotic checkpoint activation.Oncogene. 2003 Jun 19;22(25):3853-8. doi: 10.1038/sj.onc.1206502. Oncogene. 2003. PMID: 12813458
-
Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity.Mol Cancer Ther. 2002 Sep;1(11):937-41. Mol Cancer Ther. 2002. PMID: 12481415
-
Histone deacetylation is required for progression through mitosis in tobacco cells.Plant J. 2005 Feb;41(3):346-52. doi: 10.1111/j.1365-313X.2004.02301.x. Plant J. 2005. PMID: 15659094
Cited by
-
Dietary, metabolic, and potentially environmental modulation of the lysine acetylation machinery.Int J Cell Biol. 2010;2010:632739. doi: 10.1155/2010/632739. Epub 2010 Oct 5. Int J Cell Biol. 2010. PMID: 20976254 Free PMC article.
-
Anillin is an emerging regulator of tumorigenesis, acting as a cortical cytoskeletal scaffold and a nuclear modulator of cancer cell differentiation.Cell Mol Life Sci. 2021 Jan;78(2):621-633. doi: 10.1007/s00018-020-03605-9. Epub 2020 Sep 3. Cell Mol Life Sci. 2021. PMID: 32880660 Free PMC article. Review.
-
Post-translational modification of proteins in toxicological research: focus on lysine acylation.Toxicol Res. 2013 Jun;29(2):81-6. doi: 10.5487/TR.2013.29.2.081. Toxicol Res. 2013. PMID: 24278632 Free PMC article. Review.
-
An integrated overview of spatiotemporal organization and regulation in mitosis in terms of the proteins in the functional supercomplexes.Front Microbiol. 2014 Oct 29;5:573. doi: 10.3389/fmicb.2014.00573. eCollection 2014. Front Microbiol. 2014. PMID: 25400627 Free PMC article. Review.
-
RGG motif proteins: modulators of mRNA functional states.Cell Cycle. 2012 Jul 15;11(14):2594-9. doi: 10.4161/cc.20716. Epub 2012 Jul 15. Cell Cycle. 2012. PMID: 22767211 Free PMC article. Review.
References
-
- Le Breton M, Cormier P, Belle R, Mulner-Lorillon O, Morales J. Translational control during mitosis. Biochimie. 2005;87:805–811. - PubMed
-
- Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res. 2009;10:4553–4563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA090270/CA/NCI NIH HHS/United States
- P50 CA140388/CA/NCI NIH HHS/United States
- U19 AI071130-040004/AI/NIAID NIH HHS/United States
- T32 DK007696/DK/NIDDK NIH HHS/United States
- R01 CA111479-05/CA/NCI NIH HHS/United States
- S10 RR025623-01/RR/NCRR NIH HHS/United States
- RR-P20-RR17695/RR/NCRR NIH HHS/United States
- R01 DK053176-08/DK/NIDDK NIH HHS/United States
- R01 DK053176/DK/NIDDK NIH HHS/United States
- T32 DK007696-17/DK/NIDDK NIH HHS/United States
- DK53176/DK/NIDDK NIH HHS/United States
- CA111479/CA/NCI NIH HHS/United States
- AI071130/AI/NIAID NIH HHS/United States
- T32 DK007696-16/DK/NIDDK NIH HHS/United States
- SR10RR025623/RR/NCRR NIH HHS/United States
- T32 DK07696/DK/NIDDK NIH HHS/United States
- P20 RR017695/RR/NCRR NIH HHS/United States
- U19 AI071130/AI/NIAID NIH HHS/United States
- P50 CA090270/CA/NCI NIH HHS/United States
- R01 CA111479/CA/NCI NIH HHS/United States
- S10 RR025623/RR/NCRR NIH HHS/United States
- P20 RR017695-057265/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources

