Integrating common and rare genetic variation in diverse human populations
- PMID: 20811451
- PMCID: PMC3173859
- DOI: 10.1038/nature09298
Integrating common and rare genetic variation in diverse human populations
Abstract
Despite great progress in identifying genetic variants that influence human disease, most inherited risk remains unexplained. A more complete understanding requires genome-wide studies that fully examine less common alleles in populations with a wide range of ancestry. To inform the design and interpretation of such studies, we genotyped 1.6 million common single nucleotide polymorphisms (SNPs) in 1,184 reference individuals from 11 global populations, and sequenced ten 100-kilobase regions in 692 of these individuals. This integrated data set of common and rare alleles, called 'HapMap 3', includes both SNPs and copy number polymorphisms (CNPs). We characterized population-specific differences among low-frequency variants, measured the improvement in imputation accuracy afforded by the larger reference panel, especially in imputing SNPs with a minor allele frequency of <or=5%, and demonstrated the feasibility of imputing newly discovered CNPs and SNPs. This expanded public resource of genome variants in global populations supports deeper interrogation of genomic variation and its role in human disease, and serves as a step towards a high-resolution map of the landscape of human genetic variation.
Figures
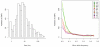
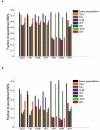
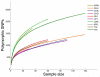
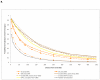

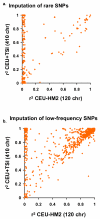
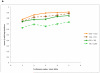

Comment in
-
Expanding HapMap.Nat Methods. 2010 Oct;7(10):780-1. doi: 10.1038/nmeth1010-780b. Nat Methods. 2010. PMID: 20936772 No abstract available.
Similar articles
-
Integrated detection and population-genetic analysis of SNPs and copy number variation.Nat Genet. 2008 Oct;40(10):1166-74. doi: 10.1038/ng.238. Epub 2008 Sep 7. Nat Genet. 2008. PMID: 18776908
-
Founder population-specific HapMap panel increases power in GWA studies through improved imputation accuracy and CNV tagging.Genome Res. 2010 Oct;20(10):1344-51. doi: 10.1101/gr.106534.110. Epub 2010 Sep 1. Genome Res. 2010. PMID: 20810666 Free PMC article.
-
A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data.J Am Med Inform Assoc. 2012 Mar-Apr;19(2):289-94. doi: 10.1136/amiajnl-2011-000652. J Am Med Inform Assoc. 2012. PMID: 22319179 Free PMC article.
-
[DNA polymorphisms].Rinsho Byori. 2013 Nov;61(11):1001-7. Rinsho Byori. 2013. PMID: 24450105 Review. Japanese.
-
Copy number variants in pharmacogenetic genes.Trends Mol Med. 2011 May;17(5):244-51. doi: 10.1016/j.molmed.2011.01.007. Epub 2011 Mar 8. Trends Mol Med. 2011. PMID: 21388883 Free PMC article. Review.
Cited by
-
Multivariate genomic analysis of 5 million people elucidates the genetic architecture of shared components of the metabolic syndrome.Nat Genet. 2024 Sep 30. doi: 10.1038/s41588-024-01933-1. Online ahead of print. Nat Genet. 2024. PMID: 39349817
-
Gene discovery and biological insights into anxiety disorders from a large-scale multi-ancestry genome-wide association study.Nat Genet. 2024 Oct;56(10):2036-2045. doi: 10.1038/s41588-024-01908-2. Epub 2024 Sep 18. Nat Genet. 2024. PMID: 39294497
-
Phenotype wide association study links bronchopulmonary dysplasia with eosinophilia in children.Sci Rep. 2024 Sep 13;14(1):21391. doi: 10.1038/s41598-024-72348-5. Sci Rep. 2024. PMID: 39271728 Free PMC article.
-
Semi-supervised machine learning method for predicting homogeneous ancestry groups to assess Hardy-Weinberg equilibrium in diverse whole-genome sequencing studies.Am J Hum Genet. 2024 Oct 3;111(10):2129-2138. doi: 10.1016/j.ajhg.2024.08.018. Epub 2024 Sep 12. Am J Hum Genet. 2024. PMID: 39270648
-
Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas.Nature. 2024 Sep;633(8029):389-397. doi: 10.1038/s41586-024-07881-4. Epub 2024 Sep 11. Nature. 2024. PMID: 39261618 Free PMC article.
References
-
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- The Internation SNP Map Working Group A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. - PubMed
-
- Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. - PubMed
Publication types
MeSH terms
Grants and funding
- P30 DK043351/DK/NIDDK NIH HHS/United States
- 068545/Z/02/WT_/Wellcome Trust/United Kingdom
- 082371/WT_/Wellcome Trust/United Kingdom
- 068545/WT_/Wellcome Trust/United Kingdom
- 077014/WT_/Wellcome Trust/United Kingdom
- G0000934/MRC_/Medical Research Council/United Kingdom
- 077011/WT_/Wellcome Trust/United Kingdom
- 076113/WT_/Wellcome Trust/United Kingdom
- U54 HG003273/HG/NHGRI NIH HHS/United States
- 091746/WT_/Wellcome Trust/United Kingdom
- 089062/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 089061/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

