Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers
- PMID: 20802487
- PMCID: PMC2957905
- DOI: 10.1038/nsmb.1891
Solid-state NMR and SAXS studies provide a structural basis for the activation of alphaB-crystallin oligomers
Abstract
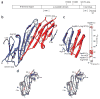
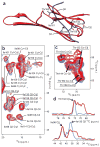
The small heat shock protein alphaB-crystallin (alphaB) contributes to cellular protection against stress. For decades, high-resolution structural studies on oligomeric alphaB have been confounded by its polydisperse nature. Here, we present a structural basis of oligomer assembly and activation of the chaperone using solid-state NMR and small-angle X-ray scattering (SAXS). The basic building block is a curved dimer, with an angle of approximately 121 degrees between the planes of the beta-sandwich formed by alpha-crystallin domains. The highly conserved IXI motif covers a substrate binding site at pH 7.5. We observe a pH-dependent modulation of the interaction of the IXI motif with beta4 and beta8, consistent with a pH-dependent regulation of the chaperone function. N-terminal region residues Ser59-Trp60-Phe61 are involved in intermolecular interaction with beta3. Intermolecular restraints from NMR and volumetric restraints from SAXS were combined to calculate a model of a 24-subunit alphaB oligomer with tetrahedral symmetry.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


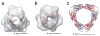
Similar articles
-
N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity.Proc Natl Acad Sci U S A. 2011 Apr 19;108(16):6409-14. doi: 10.1073/pnas.1014656108. Epub 2011 Apr 4. Proc Natl Acad Sci U S A. 2011. PMID: 21464278 Free PMC article.
-
alphaB-crystallin: a hybrid solid-state/solution-state NMR investigation reveals structural aspects of the heterogeneous oligomer.J Mol Biol. 2009 Feb 6;385(5):1481-97. doi: 10.1016/j.jmb.2008.10.097. Epub 2008 Nov 14. J Mol Biol. 2009. PMID: 19041879 Free PMC article.
-
The IXI/V motif in the C-terminal extension of alpha-crystallins: alternative interactions and oligomeric assemblies.Mol Vis. 2004 Sep 8;10:655-62. Mol Vis. 2004. PMID: 15448619
-
The multifaceted nature of αB-crystallin.Cell Stress Chaperones. 2020 Jul;25(4):639-654. doi: 10.1007/s12192-020-01098-w. Epub 2020 May 7. Cell Stress Chaperones. 2020. PMID: 32383140 Free PMC article. Review.
-
Dynamical structure of αB-crystallin.Prog Biophys Mol Biol. 2014 Jul;115(1):11-20. doi: 10.1016/j.pbiomolbio.2014.03.003. Epub 2014 Mar 24. Prog Biophys Mol Biol. 2014. PMID: 24674783 Review.
Cited by
-
The specificity of the interaction between αB-crystallin and desmin filaments and its impact on filament aggregation and cell viability.Philos Trans R Soc Lond B Biol Sci. 2013 Mar 25;368(1617):20120375. doi: 10.1098/rstb.2012.0375. Print 2013 May 5. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 23530264 Free PMC article.
-
Aging of the eye: Lessons from cataracts and age-related macular degeneration.Ageing Res Rev. 2024 Aug;99:102407. doi: 10.1016/j.arr.2024.102407. Epub 2024 Jul 6. Ageing Res Rev. 2024. PMID: 38977082 Review.
-
HSPB5 engages multiple states of a destabilized client to enhance chaperone activity in a stress-dependent manner.J Biol Chem. 2019 Mar 1;294(9):3261-3270. doi: 10.1074/jbc.RA118.003156. Epub 2018 Dec 19. J Biol Chem. 2019. PMID: 30567736 Free PMC article.
-
Effect of Structural Changes Induced by Deletion of 54FLRAPSWF61 Sequence in αB-crystallin on Chaperone Function and Anti-Apoptotic Activity.Int J Mol Sci. 2021 Oct 5;22(19):10771. doi: 10.3390/ijms221910771. Int J Mol Sci. 2021. PMID: 34639110 Free PMC article.
-
HSP27 Inhibitory Activity against Caspase-3 Cleavage and Activation by Caspase-9 Is Enhanced by Chaperone O-GlcNAc Modification in Vitro.ACS Chem Biol. 2023 Aug 18;18(8):1698-1704. doi: 10.1021/acschembio.3c00270. Epub 2023 Jul 14. ACS Chem Biol. 2023. PMID: 37450938 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

