Transcribed dark matter: meaning or myth?
- PMID: 20798109
- PMCID: PMC2953743
- DOI: 10.1093/hmg/ddq362
Transcribed dark matter: meaning or myth?
Abstract
Genomic tiling arrays, cDNA sequencing and, more recently, RNA-Seq have provided initial insights into the extent and depth of transcribed sequence across human and other genomes. These methods have led to greatly improved annotations of protein-coding genes, but have also identified transcription outside of annotated exons. One resultant issue that has aroused dispute is the balance of transcription of known exons against transcription outside of known exons. While non-genic 'dark matter' transcription was found by tiling arrays to be pervasive, it was seen to contribute only a small percentage of the polyadenylated transcriptome in some RNA-Seq experiments. This apparent contradiction has been compounded by a lack of clarity about what exactly constitutes a protein-coding gene. It remains unclear, for example, whether or not all transcripts that overlap on either strand within a genomic locus should be assigned to a single gene locus, including those that fail to share promoters, exons and splice junctions. The inability of tiling arrays and RNA-Seq to count transcripts, rather than exons or exon pairs, adds to these difficulties. While there is agreement that thousands of apparently non-coding loci are present outside of protein-coding genes in the human genome, there is vigorous debate of what constitutes evidence for their functionality. These issues will only be resolved upon the demonstration, or otherwise, that organismal or cellular phenotypes frequently result when non-coding RNA loci are disrupted.
Figures
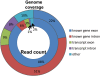

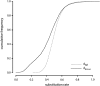
Similar articles
-
Most "dark matter" transcripts are associated with known genes.PLoS Biol. 2010 May 18;8(5):e1000371. doi: 10.1371/journal.pbio.1000371. PLoS Biol. 2010. PMID: 20502517 Free PMC article.
-
Combining RT-PCR-seq and RNA-seq to catalog all genic elements encoded in the human genome.Genome Res. 2012 Sep;22(9):1698-710. doi: 10.1101/gr.134478.111. Genome Res. 2012. PMID: 22955982 Free PMC article.
-
A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling.BMC Genomics. 2010 May 5;11:282. doi: 10.1186/1471-2164-11-282. BMC Genomics. 2010. PMID: 20444259 Free PMC article.
-
Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments.Trends Genet. 2005 Feb;21(2):93-102. doi: 10.1016/j.tig.2004.12.009. Trends Genet. 2005. PMID: 15661355 Review.
-
The Protein-Coding Human Genome: Annotating High-Hanging Fruits.Bioessays. 2019 Nov;41(11):e1900066. doi: 10.1002/bies.201900066. Epub 2019 Sep 23. Bioessays. 2019. PMID: 31544971 Review.
Cited by
-
Widespread and extensive lengthening of 3' UTRs in the mammalian brain.Genome Res. 2013 May;23(5):812-25. doi: 10.1101/gr.146886.112. Epub 2013 Mar 21. Genome Res. 2013. PMID: 23520388 Free PMC article.
-
Expression of long non-coding RNA LOC285194 and its prognostic significance in human pancreatic ductal adenocarcinoma.Int J Clin Exp Pathol. 2014 Oct 15;7(11):8065-70. eCollection 2014. Int J Clin Exp Pathol. 2014. PMID: 25550852 Free PMC article.
-
Deep analysis of wild Vitis flower transcriptome reveals unexplored genome regions associated with sex specification.Plant Mol Biol. 2017 Jan;93(1-2):151-170. doi: 10.1007/s11103-016-0553-9. Epub 2016 Oct 24. Plant Mol Biol. 2017. PMID: 27778293
-
Long noncoding RNAs: past, present, and future.Genetics. 2013 Mar;193(3):651-69. doi: 10.1534/genetics.112.146704. Genetics. 2013. PMID: 23463798 Free PMC article. Review.
-
Volatile Evolution of Long Non-Coding RNA Repertoire in Retinal Pigment Epithelium: Insights from Comparison of Bovine and Human RNA Expression Profiles.Genes (Basel). 2019 Mar 8;10(3):205. doi: 10.3390/genes10030205. Genes (Basel). 2019. PMID: 30857256 Free PMC article.
References
-
- Pertea M., Salzberg S.L. Between a chicken and a grape: estimating the number of human genes. Genome Biol. 11:206. doi:10.1186/gb-2010-11-5-206. - DOI - PMC - PubMed
-
- Ponting C.P. The functional repertoires of metazoan genomes. Nat. Rev. Genet. 2008;9:689–698. doi:10.1038/nrg2413. - DOI - PubMed
-
- Dinger M.E., Pang K.C., Mercer T.R., Mattick J.S. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput. Biol. 2008;4:e1000176. doi:10.1371/journal.pcbi.1000176. - DOI - PMC - PubMed
-
- Frith M.C., Forrest A.R., Nourbakhsh E., Pang K.C., Kai C., Kawai J., Carninci P., Hayashizaki Y., Bailey T.L., Grimmond S.M. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi:10.1371/journal.pgen.0020052. - DOI - PMC - PubMed
-
- Yamada K., Lim J., Dale J.M., Chen H., Shinn P., Palm C.J., Southwick A.M., Wu H.C., Kim C., Nguyen M., et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi:10.1126/science.1088305. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

