An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1
- PMID: 20739539
- PMCID: PMC2953177
- DOI: 10.1128/JVI.01047-10
An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1
Abstract
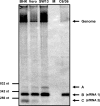
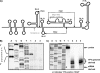
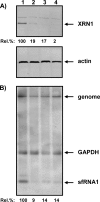
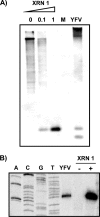
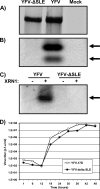
Cells and mice infected with arthropod-borne flaviviruses produce a small subgenomic RNA that is colinear with the distal part of the viral 3'-untranslated region (UTR). This small subgenomic flavivirus RNA (sfRNA) results from the incomplete degradation of the viral genome by the host 5'-3' exonuclease XRN1. Production of the sfRNA is important for the pathogenicity of the virus. This study not only presents a detailed description of the yellow fever virus (YFV) sfRNA but, more importantly, describes for the first time the molecular characteristics of the stalling site for XRN1 in the flavivirus genome. Similar to the case for West Nile virus, the YFV sfRNA was produced by XRN1. However, in contrast to the case for other arthropod-borne flaviviruses, not one but two sfRNAs were detected in YFV-infected mammalian cells. The smaller of these two sfRNAs was not observed in infected mosquito cells. The larger sfRNA could also be produced in vitro by incubation with purified XRN1. These two YFV sfRNAs formed a 5'-nested set. The 5' ends of the YFV sfRNAs were found to be just upstream of the previously predicted RNA pseudoknot PSK3. RNA structure probing and mutagenesis studies provided strong evidence that this pseudoknot structure was formed and served as the molecular signal to stall XRN1. The sequence involved in PSK3 formation was cloned into the Sinrep5 expression vector and shown to direct the production of an sfRNA-like RNA. These results underscore the importance of the RNA pseudoknot in stalling XRN1 and also demonstrate that it is the sole viral requirement for sfRNA production.
Figures

Similar articles
-
Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes.J Virol. 2016 Oct 28;90(22):10145-10159. doi: 10.1128/JVI.00930-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581979 Free PMC article.
-
Short Direct Repeats in the 3' Untranslated Region Are Involved in Subgenomic Flaviviral RNA Production.J Virol. 2020 Feb 28;94(6):e01175-19. doi: 10.1128/JVI.01175-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896596 Free PMC article.
-
Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells.J Virol. 2020 Aug 31;94(18):e00343-20. doi: 10.1128/JVI.00343-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32581095 Free PMC article.
-
Subgenomic flaviviral RNAs: What do we know after the first decade of research.Antiviral Res. 2018 Nov;159:13-25. doi: 10.1016/j.antiviral.2018.09.006. Epub 2018 Sep 11. Antiviral Res. 2018. PMID: 30217649 Review.
-
Functional non-coding RNAs derived from the flavivirus 3' untranslated region.Virus Res. 2015 Aug 3;206:53-61. doi: 10.1016/j.virusres.2015.01.026. Epub 2015 Feb 7. Virus Res. 2015. PMID: 25660582 Review.
Cited by
-
Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes.J Virol. 2016 Oct 28;90(22):10145-10159. doi: 10.1128/JVI.00930-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581979 Free PMC article.
-
The conserved stem-loop II structure at the 3' untranslated region of Japanese encephalitis virus genome is required for the formation of subgenomic flaviviral RNA.PLoS One. 2018 Jul 26;13(7):e0201250. doi: 10.1371/journal.pone.0201250. eCollection 2018. PLoS One. 2018. PMID: 30048535 Free PMC article.
-
Strategies for viral RNA stability: live long and prosper.Trends Genet. 2011 Jul;27(7):286-93. doi: 10.1016/j.tig.2011.04.003. Trends Genet. 2011. PMID: 21640425 Free PMC article. Review.
-
Musashi binding elements in Zika and related Flavivirus 3'UTRs: A comparative study in silico.Sci Rep. 2019 May 6;9(1):6911. doi: 10.1038/s41598-019-43390-5. Sci Rep. 2019. PMID: 31061405 Free PMC article.
-
Numerical integration methods and layout improvements in the context of dynamic RNA visualization.BMC Bioinformatics. 2017 May 30;18(1):282. doi: 10.1186/s12859-017-1682-0. BMC Bioinformatics. 2017. PMID: 28558664 Free PMC article.
References
-
- Alvarez, D. E., A. L. De Lella Ezcurra, S. Fucito, and A. V. Gamarnik. 2005. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 339:200-212. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. Wiley Interscience, New York, NY.
-
- Bonisch, C., C. Temme, B. Moritz, and E. Wahle. 2007. Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J. Biol. Chem. 282:21818-21828. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

