Identification of a lipid peroxidation product as the source of oxidation-specific epitopes recognized by anti-DNA autoantibodies
- PMID: 20736172
- PMCID: PMC2962483
- DOI: 10.1074/jbc.M110.165175
Identification of a lipid peroxidation product as the source of oxidation-specific epitopes recognized by anti-DNA autoantibodies
Abstract
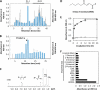
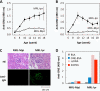
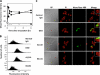
Lipid peroxidation in tissue and in tissue fractions represents a degradative process, which is the consequence of the production and the propagation of free radical reactions primarily involving membrane polyunsaturated fatty acids, and has been implicated in the pathogenesis of numerous diseases, including systemic lupus erythematosus (SLE). We have found that bovine serum albumin incubated with peroxidized polyunsaturated fatty acids significantly cross-reacted with the sera from MRL-lpr mice, a representative murine model of SLE. To identify the active substances responsible for the generation of autoantigenic epitopes recognized by the SLE sera, we performed the activity-guiding separation of a principal source from 13-hydroperoxy-9Z,11E-octadecadienoic acid and identified 4-oxo-2-nonenal (ONE), a highly reactive aldehyde originating from the peroxidation of ω6 polyunsaturated fatty acids, as the source of the autoantigenic epitopes. When the age-dependent change in the antibody titer against the ONE-modified protein was measured in the sera from MRL-lpr mice and control MRL-MpJ mice, all of the MRL-lpr mice developed an anti-ONE titer, which was comparable with the anti-DNA titer. Strikingly, a subset of the anti-DNA monoclonal antibodies generated from the SLE mice showing recognition specificity toward DNA cross-reacted with the ONE-specific epitopes. Furthermore, these dual-specific antibodies rapidly bound and internalized into living cells. These findings raised the possibility that the enhanced lipid peroxidation followed by the generation of ONE may be involved in the pathogenesis of autoimmune disorders.
Figures
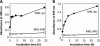

Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Medications for chronic obstructive pulmonary disease: a historical non-interventional cohort study with validation against RCT results.Health Technol Assess. 2021 Aug;25(51):1-70. doi: 10.3310/hta25510. Health Technol Assess. 2021. PMID: 34463610 Clinical Trial.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Oxidative stress as a potential causal factor for autoimmune hemolytic anemia and systemic lupus erythematosus.World J Nephrol. 2015 May 6;4(2):213-22. doi: 10.5527/wjn.v4.i2.213. World J Nephrol. 2015. PMID: 25949934 Free PMC article. Review.
-
Hypothesis: Clues From Mammalian Hibernation for Treating Patients With Anorexia Nervosa.Front Psychol. 2018 Nov 12;9:2159. doi: 10.3389/fpsyg.2018.02159. eCollection 2018. Front Psychol. 2018. PMID: 30483182 Free PMC article.
-
How does the macula protect itself from oxidative stress?Mol Aspects Med. 2012 Aug;33(4):418-35. doi: 10.1016/j.mam.2012.03.006. Epub 2012 Apr 5. Mol Aspects Med. 2012. PMID: 22503691 Free PMC article. Review.
-
Molecular and structural basis of anti-DNA antibody specificity for pyrrolated proteins.Commun Biol. 2024 Feb 3;7(1):149. doi: 10.1038/s42003-024-05851-0. Commun Biol. 2024. PMID: 38310133 Free PMC article.
-
Isolevuglandins disrupt PU.1-mediated C1q expression and promote autoimmunity and hypertension in systemic lupus erythematosus.JCI Insight. 2022 Jul 8;7(13):e136678. doi: 10.1172/jci.insight.136678. JCI Insight. 2022. PMID: 35608913 Free PMC article.
References
-
- Stadtman E. R. (1992) Science 257, 1220–1224 - PubMed
-
- Stadtman E. R., Levine R. L. (2000) Ann. N.Y. Acad. Sci. 899, 191–208 - PubMed
-
- Uchida K. (2000) Free Radic. Biol. Med. 28, 1685–1696 - PubMed
-
- Uchida K. (2003) Prog. Lipid Res. 42, 318–343 - PubMed
-
- Esterbauer H., Schaur R. J., Zollner H. (1991) Free Radic. Biol. Med. 11, 81–128 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

