The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers
- PMID: 20728025
- PMCID: PMC3073861
- DOI: 10.1016/S0065-2776(10)06004-9
The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers
Abstract
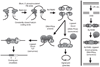
Recurrent chromosomal abnormalities, especially chromosomal translocations, are strongly associated with certain subtypes of leukemia, lymphoma and solid tumors. The appearance of particular translocations or associated genomic alterations can be important indicators of disease prognosis, and in some cases, certain translocations may indicate appropriate therapy protocols. To date, most of our knowledge about chromosomal translocations has derived from characterization of the highly selected recurrent translocations found in certain cancers. Until recently, mechanisms that promote or suppress chromosomal translocations, in particular, those responsible for their initiation, have not been addressed. For translocations to occur, two distinct chromosomal loci must be broken, brought together (synapsed) and joined. Here, we discuss recent findings on processes and pathways that influence the initiation of chromosomal translocations, including the generation fo DNA double strand breaks (DSBs) by general factors or in the context of the Lymphocyte-specific V(D)J and IgH class-switch recombination processes. We also discuss the role of spatial proximity of DSBs in the interphase nucleus with respect to how DSBs on different chromosomes are justaposed for joining. In addition, we discuss the DNA DSB response and its role in recognizing and tethering chromosomal DSBs to prevent translocations, as well as potential roles of the classical and alternative DSB end-joining pathways in suppressing or promoting translocations. Finally, we discuss the potential roles of long range regulatory elements, such as the 3'IgH enhancer complex, in promoting the expression of certain translocations that are frequent in lymphomas and, thereby, contributing to their frequent appearance in tumors.
Figures



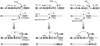
Similar articles
-
Mechanisms promoting translocations in editing and switching peripheral B cells.Nature. 2009 Jul 9;460(7252):231-6. doi: 10.1038/nature08159. Nature. 2009. PMID: 19587764 Free PMC article.
-
Mechanisms that promote and suppress chromosomal translocations in lymphocytes.Annu Rev Immunol. 2011;29:319-50. doi: 10.1146/annurev-immunol-031210-101329. Annu Rev Immunol. 2011. PMID: 21219174 Review.
-
DSB structure impacts DNA recombination leading to class switching and chromosomal translocations in human B cells.PLoS Genet. 2019 Apr 4;15(4):e1008101. doi: 10.1371/journal.pgen.1008101. eCollection 2019 Apr. PLoS Genet. 2019. PMID: 30946744 Free PMC article.
-
Developmental propagation of V(D)J recombination-associated DNA breaks and translocations in mature B cells via dicentric chromosomes.Proc Natl Acad Sci U S A. 2014 Jul 15;111(28):10269-74. doi: 10.1073/pnas.1410112111. Epub 2014 Jun 30. Proc Natl Acad Sci U S A. 2014. PMID: 24982162 Free PMC article.
-
Chromatin structural elements and chromosomal translocations in leukemia.DNA Repair (Amst). 2006 Sep 8;5(9-10):1282-97. doi: 10.1016/j.dnarep.2006.05.020. Epub 2006 Aug 7. DNA Repair (Amst). 2006. PMID: 16893685 Review.
Cited by
-
Endogenous DNA Double-Strand Breaks during DNA Transactions: Emerging Insights and Methods for Genome-Wide Profiling.Genes (Basel). 2018 Dec 14;9(12):632. doi: 10.3390/genes9120632. Genes (Basel). 2018. PMID: 30558210 Free PMC article. Review.
-
Flexible ordering of antibody class switch and V(D)J joining during B-cell ontogeny.Genes Dev. 2013 Nov 15;27(22):2439-44. doi: 10.1101/gad.227165.113. Genes Dev. 2013. PMID: 24240234 Free PMC article.
-
Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination.Cell Cycle. 2011 Jun 15;10(12):1928-35. doi: 10.4161/cc.10.12.16011. Epub 2011 Jun 15. Cell Cycle. 2011. PMID: 21673501 Free PMC article. Review.
-
Mouse model of endemic Burkitt translocations reveals the long-range boundaries of Ig-mediated oncogene deregulation.Proc Natl Acad Sci U S A. 2012 Jul 3;109(27):10972-7. doi: 10.1073/pnas.1200106109. Epub 2012 Jun 18. Proc Natl Acad Sci U S A. 2012. PMID: 22711821 Free PMC article.
-
Identification of chromosomal translocation hotspots via scan statistics.Bioinformatics. 2014 Sep 15;30(18):2551-8. doi: 10.1093/bioinformatics/btu351. Epub 2014 May 23. Bioinformatics. 2014. PMID: 24860160 Free PMC article.
References
-
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. - PubMed
-
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. - PubMed
-
- Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous endjoining. Cell. 2006;124:301–313. - PubMed
-
- Akopiant K, Zhou R-Z, Mohapatra S, Valerie K, Lees-Miller SP, Lee K-J, Chen DJ, Revy P, de Villartay J-P, Povirk LF. Requirement for XLF/Cernunnos in alignment-based gap filling by DNA polymerases l and m for nonhomolgous end joining in human whole-cell extracts. Nucleic Acids Res. 2009;37:4055–4062. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

