RNA structures required for production of subgenomic flavivirus RNA
- PMID: 20719943
- PMCID: PMC2953152
- DOI: 10.1128/JVI.01159-10
RNA structures required for production of subgenomic flavivirus RNA
Abstract
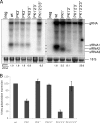
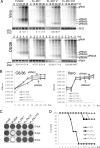
Flaviviruses are a group of single-stranded, positive-sense RNA viruses causing ∼100 million infections per year. We have recently shown that flaviviruses produce a unique, small, noncoding RNA (∼0.5 kb) derived from the 3' untranslated region (UTR) of the genomic RNA (gRNA), which is required for flavivirus-induced cytopathicity and pathogenicity (G. P. Pijlman et al., Cell Host Microbe, 4: 579-591, 2008). This RNA (subgenomic flavivirus RNA [sfRNA]) is a product of incomplete degradation of gRNA presumably by the cellular 5'-3' exoribonuclease XRN1, which stalls on the rigid secondary structure stem-loop II (SL-II) located at the beginning of the 3' UTR. Mutations or deletions of various secondary structures in the 3' UTR resulted in the loss of full-length sfRNA (sfRNA1) and production of smaller and less abundant sfRNAs (sfRNA2 and sfRNA3). Here, we investigated in detail the importance of West Nile virus Kunjin (WNV(KUN)) 3' UTR secondary structures as well as tertiary interactions for sfRNA formation. We show that secondary structures SL-IV and dumbbell 1 (DB1) downstream of SL-II are able to prevent further degradation of gRNA when the SL-II structure is deleted, leading to production of sfRNA2 and sfRNA3, respectively. We also show that a number of pseudoknot (PK) interactions, in particular PK1 stabilizing SL-II and PK3 stabilizing DB1, are required for protection of gRNA from nuclease degradation and production of sfRNA. Our results show that PK interactions play a vital role in the production of nuclease-resistant sfRNA, which is essential for viral cytopathicity in cells and pathogenicity in mice.
Figures
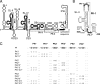
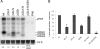
Similar articles
-
Noncoding Subgenomic Flavivirus RNA Is Processed by the Mosquito RNA Interference Machinery and Determines West Nile Virus Transmission by Culex pipiens Mosquitoes.J Virol. 2016 Oct 28;90(22):10145-10159. doi: 10.1128/JVI.00930-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581979 Free PMC article.
-
Zika Virus Subgenomic Flavivirus RNA Generation Requires Cooperativity between Duplicated RNA Structures That Are Essential for Productive Infection in Human Cells.J Virol. 2020 Aug 31;94(18):e00343-20. doi: 10.1128/JVI.00343-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32581095 Free PMC article.
-
Subgenomic flaviviral RNAs: What do we know after the first decade of research.Antiviral Res. 2018 Nov;159:13-25. doi: 10.1016/j.antiviral.2018.09.006. Epub 2018 Sep 11. Antiviral Res. 2018. PMID: 30217649 Review.
-
Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses.Viruses. 2014 Jan 27;6(2):404-27. doi: 10.3390/v6020404. Viruses. 2014. PMID: 24473339 Free PMC article. Review.
-
An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1.J Virol. 2010 Nov;84(21):11395-406. doi: 10.1128/JVI.01047-10. Epub 2010 Aug 25. J Virol. 2010. PMID: 20739539 Free PMC article.
Cited by
-
Identification of multiple RIG-I-specific pathogen associated molecular patterns within the West Nile virus genome and antigenome.Virology. 2012 Oct 10;432(1):232-8. doi: 10.1016/j.virol.2012.06.009. Epub 2012 Jul 7. Virology. 2012. PMID: 22776165 Free PMC article.
-
DDX RNA helicases: key players in cellular homeostasis and innate antiviral immunity.J Virol. 2024 Oct 22;98(10):e0004024. doi: 10.1128/jvi.00040-24. Epub 2024 Aug 30. J Virol. 2024. PMID: 39212449 Free PMC article. Review.
-
Regulation of flavivirus RNA synthesis and capping.Wiley Interdiscip Rev RNA. 2013 Nov-Dec;4(6):723-35. doi: 10.1002/wrna.1191. Epub 2013 Aug 8. Wiley Interdiscip Rev RNA. 2013. PMID: 23929625 Free PMC article. Review.
-
Functional RNA elements in the dengue virus genome.Viruses. 2011 Sep;3(9):1739-56. doi: 10.3390/v3091739. Epub 2011 Sep 15. Viruses. 2011. PMID: 21994804 Free PMC article. Review.
-
The role of innate immunity in conditioning mosquito susceptibility to West Nile virus.Viruses. 2013 Dec 13;5(12):3142-70. doi: 10.3390/v5123142. Viruses. 2013. PMID: 24351797 Free PMC article. Review.
References
-
- Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huijkman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. - PubMed
-
- Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153:113-121. - PubMed
-
- Brion, P., and E. Westhof. 1997. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26:113-137. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

