p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy
- PMID: 20716666
- PMCID: PMC2945185
- DOI: 10.1101/gr.103945.109
p53 binds preferentially to genomic regions with high DNA-encoded nucleosome occupancy
Abstract
The human transcription factor TP53 is a pivotal roadblock against cancer. A key unresolved question is how the p53 protein selects its genomic binding sites in vivo out of a large pool of potential consensus sites. We hypothesized that chromatin may play a significant role in this site-selection process. To test this, we used a custom DNA microarray to measure p53 binding at approximately 2000 sites predicted to possess high-sequence specificity, and identified both strongly bound and weakly bound sites. When placed within a plasmid, weakly bound sites become p53 responsive and regain p53 binding when stably integrated into random genomic locations. Notably, strongly bound sites reside preferentially within genomic regions whose DNA sequence is predicted to encode relatively high intrinsic nucleosome occupancy. Using in vivo nucleosome occupancy measurements under conditions where p53 is inactive, we experimentally confirmed this prediction. Furthermore, upon p53 activation, nucleosomes are partially displaced from a relatively broad region surrounding the bound p53 sites, and this displacement is rapidly reversed upon inactivation of p53. Thus, in contrast to the general assumption that transcription-factor binding is preferred in sites that have low nucleosome occupancy prior to factor activation, we find that p53 binding occurs preferentially within a chromatin context of high intrinsic nucleosome occupancy.
Figures
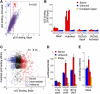
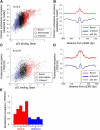
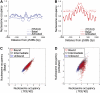

Similar articles
-
Blurring of high-resolution data shows that the effect of intrinsic nucleosome occupancy on transcription factor binding is mostly regional, not local.PLoS Comput Biol. 2010 Jan 22;6(1):e1000649. doi: 10.1371/journal.pcbi.1000649. PLoS Comput Biol. 2010. PMID: 20098497 Free PMC article.
-
Multiple distinct stimuli increase measured nucleosome occupancy around human promoters.PLoS One. 2011;6(8):e23490. doi: 10.1371/journal.pone.0023490. Epub 2011 Aug 11. PLoS One. 2011. PMID: 21853138 Free PMC article.
-
p53 binding sites in normal and cancer cells are characterized by distinct chromatin context.Cell Cycle. 2017;16(21):2073-2085. doi: 10.1080/15384101.2017.1361064. Epub 2017 Sep 28. Cell Cycle. 2017. PMID: 28820292 Free PMC article.
-
Interactions of chromatin context, binding site sequence content, and sequence evolution in stress-induced p53 occupancy and transactivation.PLoS Genet. 2015 Jan 8;11(1):e1004885. doi: 10.1371/journal.pgen.1004885. eCollection 2015 Jan. PLoS Genet. 2015. PMID: 25569532 Free PMC article.
-
Determinants of p53 DNA binding, gene regulation, and cell fate decisions.Cell Death Differ. 2024 Jul;31(7):836-843. doi: 10.1038/s41418-024-01326-1. Epub 2024 Jun 29. Cell Death Differ. 2024. PMID: 38951700 Free PMC article. Review.
Cited by
-
Structural basis for p53 binding to its nucleosomal target DNA sequence.PNAS Nexus. 2022 Sep 4;1(4):pgac177. doi: 10.1093/pnasnexus/pgac177. eCollection 2022 Sep. PNAS Nexus. 2022. PMID: 36714865 Free PMC article.
-
Preferential binding of hot spot mutant p53 proteins to supercoiled DNA in vitro and in cells.PLoS One. 2013;8(3):e59567. doi: 10.1371/journal.pone.0059567. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555710 Free PMC article.
-
Differential Nucleosome Occupancies across Oct4-Sox2 Binding Sites in Murine Embryonic Stem Cells.PLoS One. 2015 May 18;10(5):e0127214. doi: 10.1371/journal.pone.0127214. eCollection 2015. PLoS One. 2015. PMID: 25992972 Free PMC article.
-
Pioneer Factors in Animals and Plants-Colonizing Chromatin for Gene Regulation.Molecules. 2018 Jul 31;23(8):1914. doi: 10.3390/molecules23081914. Molecules. 2018. PMID: 30065231 Free PMC article. Review.
-
Transcription factor networks in erythroid cell and megakaryocyte development.Blood. 2011 Jul 14;118(2):231-9. doi: 10.1182/blood-2011-04-285981. Epub 2011 May 26. Blood. 2011. PMID: 21622645 Free PMC article. Review.
References
-
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446: 572–576 - PubMed
-
- Aylon Y, Oren M 2007. Living with p53, dying of p53. Cell 130: 597–600 - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Brummelkamp TR, Bernards R, Agami R 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 - PubMed
-
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116: 499–509 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
