Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster
- PMID: 20713603
- PMCID: PMC2928026
- DOI: 10.1083/jcb.201003001
Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster
Abstract
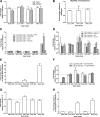
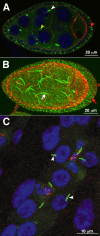
The discovery of large supramolecular complexes such as the purinosome suggests that subcellular organization is central to enzyme regulation. A screen of the yeast GFP strain collection to identify proteins that assemble into visible structures identified four novel filament systems comprised of glutamate synthase, guanosine diphosphate-mannose pyrophosphorylase, cytidine triphosphate (CTP) synthase, or subunits of the eIF2/2B translation factor complex. Recruitment of CTP synthase to filaments and foci can be modulated by mutations and regulatory ligands that alter enzyme activity, arguing that the assembly of these structures is related to control of CTP synthase activity. CTP synthase filaments are evolutionarily conserved and are restricted to axons in neurons. This spatial regulation suggests that these filaments have additional functions separate from the regulation of enzyme activity. The identification of four novel filaments greatly expands the number of known intracellular filament networks and has broad implications for our understanding of how cells organize biochemical activities in the cytoplasm.
Figures


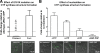
Similar articles
-
Common regulatory control of CTP synthase enzyme activity and filament formation.Mol Biol Cell. 2014 Aug 1;25(15):2282-90. doi: 10.1091/mbc.E14-04-0912. Epub 2014 Jun 11. Mol Biol Cell. 2014. PMID: 24920825 Free PMC article.
-
Characterization of filament-forming CTP synthases from Arabidopsis thaliana.Plant J. 2018 Oct;96(2):316-328. doi: 10.1111/tpj.14032. Epub 2018 Aug 31. Plant J. 2018. PMID: 30030857 Free PMC article.
-
Intracellular compartmentation of CTP synthase in Drosophila.J Genet Genomics. 2010 May;37(5):281-96. doi: 10.1016/S1673-8527(09)60046-1. J Genet Genomics. 2010. PMID: 20513629
-
Phospholipid synthesis in yeast: regulation by phosphorylation.Biochem Cell Biol. 2004 Feb;82(1):62-70. doi: 10.1139/o03-064. Biochem Cell Biol. 2004. PMID: 15052328 Review.
-
The GAL4 System: A Versatile System for the Manipulation and Analysis of Gene Expression.Methods Mol Biol. 2016;1478:33-52. doi: 10.1007/978-1-4939-6371-3_2. Methods Mol Biol. 2016. PMID: 27730574 Review.
Cited by
-
Protein aggregation as a mechanism of adaptive cellular responses.Curr Genet. 2016 Nov;62(4):711-724. doi: 10.1007/s00294-016-0596-0. Epub 2016 Mar 31. Curr Genet. 2016. PMID: 27032776 Review.
-
A quantitative screen for metabolic enzyme structures reveals patterns of assembly across the yeast metabolic network.Mol Biol Cell. 2019 Oct 1;30(21):2721-2736. doi: 10.1091/mbc.E19-04-0224. Epub 2019 Sep 4. Mol Biol Cell. 2019. PMID: 31483745 Free PMC article.
-
Saccharomyces cerevisiae ASN1 and ASN2 are asparagine synthetase paralogs that have diverged in their ability to polymerize in response to nutrient stress.Sci Rep. 2019 Jan 22;9(1):278. doi: 10.1038/s41598-018-36719-z. Sci Rep. 2019. PMID: 30670751 Free PMC article.
-
An array of nuclear microtubules reorganizes the budding yeast nucleus during quiescence.J Cell Biol. 2013 Nov 25;203(4):585-94. doi: 10.1083/jcb.201306075. Epub 2013 Nov 18. J Cell Biol. 2013. PMID: 24247429 Free PMC article.
-
An aggregation-prone mutant of eIF3a forms reversible assemblies escaping spatial control in exponentially growing yeast cells.Curr Genet. 2019 Aug;65(4):919-940. doi: 10.1007/s00294-019-00940-8. Epub 2019 Feb 4. Curr Genet. 2019. PMID: 30715564
References
-
- Aronow B., Ullman B. 1987. In situ regulation of mammalian CTP synthetase by allosteric inhibition. J. Biol. Chem. 262:5106–5112 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

