Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish
- PMID: 20707909
- PMCID: PMC3091668
- DOI: 10.1186/1471-2164-11-472
Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish
Abstract
Background: Systematic research on fish immunogenetics is indispensable in understanding the origin and evolution of immune systems. This has long been a challenging task because of the limited number of deep sequencing technologies and genome backgrounds of non-model fish available. The newly developed Solexa/Illumina RNA-seq and Digital gene expression (DGE) are high-throughput sequencing approaches and are powerful tools for genomic studies at the transcriptome level. This study reports the transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus using RNA-seq and DGE in an attempt to gain insights into the immunogenetics of marine fish.
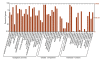
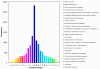
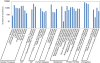
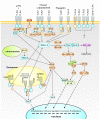
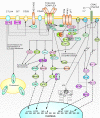
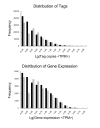
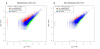
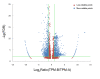
Results: RNA-seq analysis generated 169,950 non-redundant consensus sequences, among which 48,987 functional transcripts with complete or various length encoding regions were identified. More than 52% of these transcripts are possibly involved in approximately 219 known metabolic or signalling pathways, while 2,673 transcripts were associated with immune-relevant genes. In addition, approximately 8% of the transcripts appeared to be fish-specific genes that have never been described before. DGE analysis revealed that the host transcriptome profile of Vibrio harveyi-challenged L. japonicus is considerably altered, as indicated by the significant up- or down-regulation of 1,224 strong infection-responsive transcripts. Results indicated an overall conservation of the components and transcriptome alterations underlying innate and adaptive immunity in fish and other vertebrate models. Analysis suggested the acquisition of numerous fish-specific immune system components during early vertebrate evolution.
Conclusion: This study provided a global survey of host defence gene activities against bacterial challenge in a non-model marine fish. Results can contribute to the in-depth study of candidate genes in marine fish immunity, and help improve current understanding of host-pathogen interactions and evolutionary history of immunogenetics from fish to mammals.
Figures
Similar articles
-
RNA-Seq analysis of immune-relevant genes in Lateolabrax japonicus during Vibrio anguillarum infection.Fish Shellfish Immunol. 2016 May;52:57-64. doi: 10.1016/j.fsi.2016.02.032. Epub 2016 Mar 2. Fish Shellfish Immunol. 2016. PMID: 26945936
-
Analysis of apolipoprotein multigene family in spotted sea bass (Lateolabrax maculatus) and their expression profiles in response to Vibrio harveyi infection.Fish Shellfish Immunol. 2019 Sep;92:111-118. doi: 10.1016/j.fsi.2019.06.005. Epub 2019 Jun 6. Fish Shellfish Immunol. 2019. PMID: 31176005
-
The dynamic immune response of the liver and spleen in leopard coral grouper (Plectropomus leopardus) to Vibrio harveyi infection based on transcriptome analysis.Front Immunol. 2024 Oct 10;15:1457745. doi: 10.3389/fimmu.2024.1457745. eCollection 2024. Front Immunol. 2024. PMID: 39450165 Free PMC article.
-
Comparative Study of Immune Reaction Against Bacterial Infection From Transcriptome Analysis.Front Immunol. 2019 Feb 5;10:153. doi: 10.3389/fimmu.2019.00153. eCollection 2019. Front Immunol. 2019. PMID: 30804945 Free PMC article. Review.
-
Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review.Int J Mol Sci. 2018 Jan 15;19(1):245. doi: 10.3390/ijms19010245. Int J Mol Sci. 2018. PMID: 29342931 Free PMC article. Review.
Cited by
-
Transcriptome analysis of Nicotiana tabacum infected by Cucumber mosaic virus during systemic symptom development.PLoS One. 2012;7(8):e43447. doi: 10.1371/journal.pone.0043447. Epub 2012 Aug 28. PLoS One. 2012. PMID: 22952684 Free PMC article.
-
Transcriptome of Atlantic cod (Gadus morhua L.) early embryos from farmed and wild broodstocks.Mar Biotechnol (NY). 2013 Dec;15(6):677-94. doi: 10.1007/s10126-013-9527-y. Epub 2013 Jul 27. Mar Biotechnol (NY). 2013. PMID: 23887676
-
Applications for next-generation sequencing in fish ecotoxicogenomics.Front Genet. 2012 Apr 25;3:62. doi: 10.3389/fgene.2012.00062. eCollection 2012. Front Genet. 2012. PMID: 22539934 Free PMC article.
-
Transcriptome Comparison Analysis of Ostrinia furnacalis in Four Developmental Stages.Sci Rep. 2016 Oct 7;6:35008. doi: 10.1038/srep35008. Sci Rep. 2016. PMID: 27713521 Free PMC article.
-
Maternal control of seed weight in rapeseed (Brassica napus L.): the causal link between the size of pod (mother, source) and seed (offspring, sink).Plant Biotechnol J. 2019 Apr;17(4):736-749. doi: 10.1111/pbi.13011. Epub 2018 Nov 28. Plant Biotechnol J. 2019. PMID: 30191657 Free PMC article.
References
-
- Zou J, Tafalla C, Truckle J, Secombes CJ. Identification of a second group of type I IFNs in fish sheds light on IFN evolution in vertebrates. J Immunol. 2007;179(6):3859–3871. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

