The ESEV PDZ-binding motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis by directly targeting Scribble
- PMID: 20702615
- PMCID: PMC2953166
- DOI: 10.1128/JVI.01278-10
The ESEV PDZ-binding motif of the avian influenza A virus NS1 protein protects infected cells from apoptosis by directly targeting Scribble
Abstract
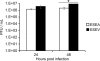
The NS1 protein from influenza A viruses contains a four-amino-acid sequence at its carboxyl terminus that is termed the PDZ-binding motif (PBM). The NS1 PBM is predicted to bind to cellular PDZ proteins and functions as a virulence determinant in infected mice. ESEV is the consensus PBM sequence of avian influenza viruses, while RSKV is the consensus sequence of human viruses. Currently circulating highly pathogenic H5N1 influenza viruses encode an NS1 protein with the ESEV PBM. We identified cellular targets of the avian ESEV PBM and identified molecular mechanisms involved in its function. Using glutathione S-transferase (GST) pull-down assays, we found that the ESEV PBM enables NS1 to associate with the PDZ proteins Scribble, Dlg1, MAGI-1, MAGI-2, and MAGI-3. Because Scribble possesses a proapoptotic activity, we investigated the interaction between NS1 and Scribble. The association between NS1 and Scribble is direct and requires the ESEV PBM and two Scribble PDZ domains. We constructed recombinant H3N2 viruses that encode an H6N6 avian virus NS1 protein with either an ESEV or mutant ESEA PBM, allowing an analysis of the ESEV PBM in infections in mammalian cells. The ESEV PBM enhanced viral replication up to 4-fold. In infected cells, NS1 with the ESEV PBM relocalized Scribble into cytoplasmic puncta concentrated in perinuclear regions and also protected cells from apoptosis. In addition, the latter effect was eliminated by small interfering RNA (siRNA)-mediated Scribble depletion. This study shows that one function of the avian ESEV PBM is to reduce apoptosis during infection through disruption of Scribble's proapoptotic function.
Figures


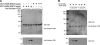

Similar articles
-
The avian influenza virus NS1 ESEV PDZ binding motif associates with Dlg1 and Scribble to disrupt cellular tight junctions.J Virol. 2011 Oct;85(20):10639-48. doi: 10.1128/JVI.05070-11. Epub 2011 Aug 17. J Virol. 2011. PMID: 21849460 Free PMC article.
-
Regulation of interferon-β by MAGI-1 and its interaction with influenza A virus NS1 protein with ESEV PBM.PLoS One. 2012;7(7):e41251. doi: 10.1371/journal.pone.0041251. Epub 2012 Jul 20. PLoS One. 2012. PMID: 22911767 Free PMC article.
-
PDlim2 selectively interacts with the PDZ binding motif of highly pathogenic avian H5N1 influenza A virus NS1.PLoS One. 2011;6(5):e19511. doi: 10.1371/journal.pone.0019511. Epub 2011 May 23. PLoS One. 2011. PMID: 21625420 Free PMC article.
-
Structure and function of the NS1 protein of influenza A virus.Acta Biochim Biophys Sin (Shanghai). 2007 Mar;39(3):155-62. doi: 10.1111/j.1745-7270.2007.00263.x. Acta Biochim Biophys Sin (Shanghai). 2007. PMID: 17342253 Review.
-
Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses.J Virol. 2011 Nov;85(22):11544-56. doi: 10.1128/JVI.05410-11. Epub 2011 Jul 20. J Virol. 2011. PMID: 21775458 Free PMC article. Review.
Cited by
-
The Cellular DExD/H-Box RNA Helicase UAP56 Co-localizes With the Influenza A Virus NS1 Protein.Front Microbiol. 2018 Sep 12;9:2192. doi: 10.3389/fmicb.2018.02192. eCollection 2018. Front Microbiol. 2018. PMID: 30258431 Free PMC article.
-
A comprehensive map of the influenza A virus replication cycle.BMC Syst Biol. 2013 Oct 2;7:97. doi: 10.1186/1752-0509-7-97. BMC Syst Biol. 2013. PMID: 24088197 Free PMC article.
-
Comprehensive proteomic analysis of influenza virus polymerase complex reveals a novel association with mitochondrial proteins and RNA polymerase accessory factors.J Virol. 2011 Sep;85(17):8569-81. doi: 10.1128/JVI.00496-11. Epub 2011 Jun 29. J Virol. 2011. PMID: 21715506 Free PMC article.
-
The NS1 protein of influenza A virus suppresses interferon-regulated activation of antigen-presentation and immune-proteasome pathways.J Gen Virol. 2011 Sep;92(Pt 9):2093-2104. doi: 10.1099/vir.0.032060-0. Epub 2011 May 18. J Gen Virol. 2011. PMID: 21593271 Free PMC article.
-
Viral Interactions with PDZ Domain-Containing Proteins-An Oncogenic Trait?Pathogens. 2016 Jan 18;5(1):8. doi: 10.3390/pathogens5010008. Pathogens. 2016. PMID: 26797638 Free PMC article. Review.
References
-
- Arpin-Andre, C., and J. M. Mesnard. 2007. The PDZ domain-binding motif of the human T cell leukemia virus type 1 tax protein induces mislocalization of the tumor suppressor hScrib in T cells. J. Biol. Chem. 282:33132-33141. - PubMed
-
- Assemat, E., E. Bazellieres, E. Pallesi-Pocachard, A. Le Bivic, and D. Massey-Harroche. 2008. Polarity complex proteins. Biochim. Biophys. Acta 1778:614-630. - PubMed
-
- Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. - PubMed
-
- Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

