Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes
- PMID: 20691578
- PMCID: PMC3008753
- DOI: 10.1016/j.jnutbio.2010.02.011
Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes
Abstract
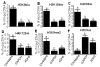
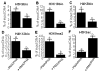
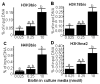
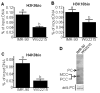
Covalent histone modifications play crucial roles in chromatin structure and genome stability. We previously reported biotinylation of lysine (K) residues in histones H2A, H3 and H4 by holocarboxylase synthetase and demonstrated that K12-biotinylated histone H4 (H4K12bio) is enriched in repeat regions and participates in gene repression. The biological functions of biotinylation marks other than H4K12bio are poorly understood. Here, novel biotinylation site-specific antibodies against H3K9bio, H3K18bio and H4K8bio were used in chromatin immunoprecipitation studies to obtain first insights into possible biological functions of these marks. Chromatin immunoprecipitation assays were conducted in human primary fibroblasts and Jurkat lymphoblastoma cells, and revealed that H3K9bio, H3K18bio and H4K8bio are enriched in repeat regions such as pericentromeric alpha satellite repeats and long-terminal repeats while being depleted in transcriptionally active promoters in euchromatin. Transcriptional stimulation of the repressed interleukin-2 promoter triggered a rapid depletion of histone biotinylation marks at this locus in Jurkat cells, which was paralleled by an increase in interleukin-2 mRNA. Importantly, the enrichment of H3K9bio, H3K18bio and H4K8bio at genomic loci depended on the concentration of biotin in culture media at nutritionally relevant levels, suggesting a novel mechanism of gene regulation by biotin.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
K16-biotinylated histone H4 is overrepresented in repeat regions and participates in the repression of transcriptionally competent genes in human Jurkat lymphoid cells.J Nutr Biochem. 2012 Dec;23(12):1559-64. doi: 10.1016/j.jnutbio.2011.10.009. Epub 2011 Dec 20. J Nutr Biochem. 2012. PMID: 22192339 Free PMC article.
-
K12-biotinylated histone H4 marks heterochromatin in human lymphoblastoma cells.J Nutr Biochem. 2007 Nov;18(11):760-8. doi: 10.1016/j.jnutbio.2006.12.014. Epub 2007 Apr 16. J Nutr Biochem. 2007. PMID: 17434721 Free PMC article.
-
K12-biotinylated histone H4 is enriched in telomeric repeats from human lung IMR-90 fibroblasts.J Nutr Biochem. 2010 Apr;21(4):310-6. doi: 10.1016/j.jnutbio.2009.01.010. Epub 2009 Apr 14. J Nutr Biochem. 2010. PMID: 19369050 Free PMC article.
-
A novel, enigmatic histone modification: biotinylation of histones by holocarboxylase synthetase.Nutr Rev. 2008 Dec;66(12):721-5. doi: 10.1111/j.1753-4887.2008.00127.x. Nutr Rev. 2008. PMID: 19019041 Review.
-
Epigenetic regulation of chromatin structure and gene function by biotin.J Nutr. 2006 Jul;136(7):1763-5. doi: 10.1093/jn/136.7.1763. J Nutr. 2006. PMID: 16772434 Free PMC article. Review.
Cited by
-
Holocarboxylase synthetase interacts physically with euchromatic histone-lysine N-methyltransferase, linking histone biotinylation with methylation events.J Nutr Biochem. 2013 Aug;24(8):1446-52. doi: 10.1016/j.jnutbio.2012.12.003. Epub 2013 Jan 20. J Nutr Biochem. 2013. PMID: 23337344 Free PMC article.
-
Epigenetic synergies between biotin and folate in the regulation of pro-inflammatory cytokines and repeats.Scand J Immunol. 2013 Nov;78(5):419-25. doi: 10.1111/sji.12108. Scand J Immunol. 2013. PMID: 24007195 Free PMC article.
-
Transcriptional regulation of the albumin gene depends on the removal of histone methylation marks by the FAD-dependent monoamine oxidase lysine-specific demethylase 1 in HepG2 human hepatocarcinoma cells.J Nutr. 2014 Jul;144(7):997-1001. doi: 10.3945/jn.114.192187. Epub 2014 Apr 17. J Nutr. 2014. PMID: 24744315 Free PMC article.
-
A diet defined by its content of bovine milk exosomes and their RNA cargos has moderate effects on gene expression, amino acid profiles and grip strength in skeletal muscle in C57BL/6 mice.J Nutr Biochem. 2018 Sep;59:123-128. doi: 10.1016/j.jnutbio.2018.06.007. Epub 2018 Jun 12. J Nutr Biochem. 2018. PMID: 29986306 Free PMC article.
-
The Potential Role of Gut Microbiota in the Pathogenesis of Type 2 Diabetes Mellitus via Epigenetics and Inflammasome.Endocr Metab Immune Disord Drug Targets. 2022;22(14):1331-1343. doi: 10.2174/1871530322666220331152809. Endocr Metab Immune Disord Drug Targets. 2022. PMID: 35362379 Review.
References
-
- Wolffe A. Chromatin. 3. Academic Press; San Diego, CA: 1998.
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–60. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. - PubMed
-
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. - PubMed
-
- Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 DK082476/DK/NIDDK NIH HHS/United States
- DK077816/DK/NIDDK NIH HHS/United States
- ES015206/ES/NIEHS NIH HHS/United States
- R01 DK063945/DK/NIDDK NIH HHS/United States
- R21 ES015206/ES/NIEHS NIH HHS/United States
- R01 DK077816-01A2S1/DK/NIDDK NIH HHS/United States
- R01 DK063945-06/DK/NIDDK NIH HHS/United States
- DK082476/DK/NIDDK NIH HHS/United States
- DK063945/DK/NIDDK NIH HHS/United States
- R21 DK082476-01/DK/NIDDK NIH HHS/United States
- R01 DK077816-02/DK/NIDDK NIH HHS/United States
- R01 DK077816/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources

