Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis
- PMID: 20687163
- PMCID: PMC2998727
- DOI: 10.1002/pro.480
Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis
Abstract
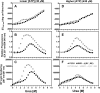
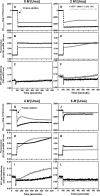
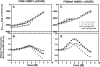
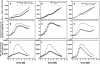
The lethal genetic disease cystic fibrosis is caused predominantly by in-frame deletion of phenylalanine 508 in the cystic fibrosis transmembrane conductance regulator (CFTR). F508 is located in the first nucleotide-binding domain (NBD1) of CFTR, which functions as an ATP-gated chloride channel on the cell surface. The F508del mutation blocks CFTR export to the surface due to aberrant retention in the endoplasmic reticulum. While it was assumed that F508del interferes with NBD1 folding, biophysical studies of purified NBD1 have given conflicting results concerning the mutation's influence on domain folding and stability. We have conducted isothermal (this paper) and thermal (accompanying paper) denaturation studies of human NBD1 using a variety of biophysical techniques, including simultaneous circular dichroism, intrinsic fluorescence, and static light-scattering measurements. These studies show that, in the absence of ATP, NBD1 unfolds via two sequential conformational transitions. The first, which is strongly influenced by F508del, involves partial unfolding and leads to aggregation accompanied by an increase in tryptophan fluorescence. The second, which is not significantly influenced by F508del, involves full unfolding of NBD1. Mg-ATP binding delays the first transition, thereby offsetting the effect of F508del on domain stability. Evidence suggests that the initial partial unfolding transition is partially responsible for the poor in vitro solubility of human NBD1. Second-site mutations that increase the solubility of isolated F508del-NBD1 in vitro and suppress the trafficking defect of intact F508del-CFTR in vivo also stabilize the protein against this transition, supporting the hypothesize that it is responsible for the pathological trafficking of F508del-CFTR.
Figures

Similar articles
-
Dynamics intrinsic to cystic fibrosis transmembrane conductance regulator function and stability.Cold Spring Harb Perspect Med. 2013 Mar 1;3(3):a009522. doi: 10.1101/cshperspect.a009522. Cold Spring Harb Perspect Med. 2013. PMID: 23457292 Free PMC article. Review.
-
Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1.Protein Sci. 2010 Oct;19(10):1917-31. doi: 10.1002/pro.479. Protein Sci. 2010. PMID: 20687133 Free PMC article.
-
Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure.J Biol Chem. 2005 Jan 14;280(2):1346-53. doi: 10.1074/jbc.M410968200. Epub 2004 Nov 3. J Biol Chem. 2005. PMID: 15528182
-
Non-native Conformers of Cystic Fibrosis Transmembrane Conductance Regulator NBD1 Are Recognized by Hsp27 and Conjugated to SUMO-2 for Degradation.J Biol Chem. 2016 Jan 22;291(4):2004-2017. doi: 10.1074/jbc.M115.685628. Epub 2015 Dec 1. J Biol Chem. 2016. PMID: 26627832 Free PMC article.
-
Decoding F508del misfolding in cystic fibrosis.Biomolecules. 2014 May 6;4(2):498-509. doi: 10.3390/biom4020498. Biomolecules. 2014. PMID: 24970227 Free PMC article. Review.
Cited by
-
Dynamics intrinsic to cystic fibrosis transmembrane conductance regulator function and stability.Cold Spring Harb Perspect Med. 2013 Mar 1;3(3):a009522. doi: 10.1101/cshperspect.a009522. Cold Spring Harb Perspect Med. 2013. PMID: 23457292 Free PMC article. Review.
-
Allosteric modulation balances thermodynamic stability and restores function of ΔF508 CFTR.J Mol Biol. 2012 May 25;419(1-2):41-60. doi: 10.1016/j.jmb.2012.03.001. Epub 2012 Mar 8. J Mol Biol. 2012. PMID: 22406676 Free PMC article.
-
Rational Coupled Dynamics Network Manipulation Rescues Disease-Relevant Mutant Cystic Fibrosis Transmembrane Conductance Regulator.Chem Sci. 2015 Feb;6(2):1237-1246. doi: 10.1039/C4SC01320D. Chem Sci. 2015. PMID: 25685315 Free PMC article.
-
Molecular modelling and molecular dynamics of CFTR.Cell Mol Life Sci. 2017 Jan;74(1):3-22. doi: 10.1007/s00018-016-2385-9. Epub 2016 Oct 7. Cell Mol Life Sci. 2017. PMID: 27717958 Free PMC article. Review.
-
Disruption of cytokeratin-8 interaction with F508del-CFTR corrects its functional defect.Hum Mol Genet. 2012 Feb 1;21(3):623-34. doi: 10.1093/hmg/ddr496. Epub 2011 Oct 28. Hum Mol Genet. 2012. PMID: 22038833 Free PMC article.
References
-
- Riordan JR, Rommens JM, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou JL, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. - PubMed
-
- Anderson MP, Gregory RJ, Thompson S, Souza DW, Paul S, Mulligan RC, Smith AE, Welsh MJ. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science. 1991;253:202–205. - PubMed
-
- Anderson MP, Rich DP, Gregory RJ, Smith AE, Welsh MJ. Generation of cAMP-activated chloride currents by expression of CFTR. Science. 1991;251:679–682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

