Interferon-gamma-mediated inhibition of serum response factor-dependent smooth muscle-specific gene expression
- PMID: 20685657
- PMCID: PMC2952243
- DOI: 10.1074/jbc.M110.164863
Interferon-gamma-mediated inhibition of serum response factor-dependent smooth muscle-specific gene expression
Abstract
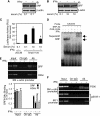
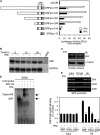
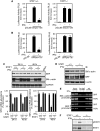
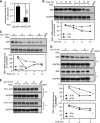
IFNγ exerts multiple biological effects on effector cells by regulating many downstream genes, including smooth muscle-specific genes. However, the molecular mechanisms underlying IFNγ-induced inhibition of smooth muscle-specific gene expression remain unclear. In this study, we have shown that serum response factor (SRF), a common transcriptional factor important in cell proliferation, migration, and differentiation, is targeted by IFNγ in a STAT1-dependent manner. We show that the molecular mechanism by which IFNγ regulates SRF is via activation of the 2-5A-RNase L system, which triggers SRF mRNA decay and reduced SRF expression. As a result, decreased SRF expression reduces expression of SRF target genes such as smooth muscle α-actin and smooth muscle myosin heavy chain. Additionally, IFNγ reduced p300 and acetylated histone-3 binding in both smooth muscle α-actin and SRF promoters, epigenetically decreasing smooth muscle α-actin and SRF transcriptional activation. Our data reveal that SRF is a novel IFNγ-regulated gene and further elucidate the molecular pathway between IFNγ, IFNγ-regulated genes, and SRF and its target genes.
Figures


Similar articles
-
The RhoA/Rho kinase pathway regulates nuclear localization of serum response factor.Am J Respir Cell Mol Biol. 2003 Jul;29(1):39-47. doi: 10.1165/rcmb.2002-0206OC. Epub 2003 Jan 10. Am J Respir Cell Mol Biol. 2003. PMID: 12600823
-
Ablation of serum response factor in hepatic stellate cells attenuates liver fibrosis.J Mol Med (Berl). 2019 Nov;97(11):1521-1533. doi: 10.1007/s00109-019-01831-8. Epub 2019 Aug 21. J Mol Med (Berl). 2019. PMID: 31435710
-
Serum response factor neutralizes Pur alpha- and Pur beta-mediated repression of the fetal vascular smooth muscle alpha-actin gene in stressed adult cardiomyocytes.Am J Physiol Cell Physiol. 2008 Mar;294(3):C702-14. doi: 10.1152/ajpcell.00173.2007. Epub 2008 Jan 9. Am J Physiol Cell Physiol. 2008. PMID: 18344281
-
Serum response factor: toggling between disparate programs of gene expression.J Mol Cell Cardiol. 2003 Jun;35(6):577-93. doi: 10.1016/s0022-2828(03)00110-x. J Mol Cell Cardiol. 2003. PMID: 12788374 Review.
-
SRF in Neurochemistry: Overview of Recent Advances in Research on the Nervous System.Neurochem Res. 2022 Sep;47(9):2545-2557. doi: 10.1007/s11064-022-03632-x. Epub 2022 Jun 7. Neurochem Res. 2022. PMID: 35668335 Review.
Cited by
-
Interferon-γ blocks signalling through PDGFRβ in human brain pericytes.J Neuroinflammation. 2016 Sep 21;13(1):249. doi: 10.1186/s12974-016-0722-4. J Neuroinflammation. 2016. PMID: 27654972 Free PMC article.
-
Caveolin-1 is upregulated in hepatic stellate cells but not sinusoidal endothelial cells after liver injury.Tissue Cell. 2016 Apr;48(2):126-32. doi: 10.1016/j.tice.2015.12.006. Epub 2016 Jan 4. Tissue Cell. 2016. PMID: 26847875 Free PMC article.
-
The mitochondria-targeted antioxidant MitoQ attenuates liver fibrosis in mice.Int J Physiol Pathophysiol Pharmacol. 2016 Apr 25;8(1):14-27. eCollection 2016. Int J Physiol Pathophysiol Pharmacol. 2016. PMID: 27186319 Free PMC article.
-
Fibroblast-specific Stat1 deletion enhances the myofibroblast phenotype during tissue repair.Wound Repair Regen. 2020 Jul;28(4):448-459. doi: 10.1111/wrr.12807. Epub 2020 Mar 24. Wound Repair Regen. 2020. PMID: 32175700 Free PMC article.
-
Smooth Muscle α-Actin Deficiency Leads to Decreased Liver Fibrosis via Impaired Cytoskeletal Signaling in Hepatic Stellate Cells.Am J Pathol. 2019 Nov;189(11):2209-2220. doi: 10.1016/j.ajpath.2019.07.019. Epub 2019 Aug 30. Am J Pathol. 2019. PMID: 31476284 Free PMC article.
References
-
- Boehm U., Klamp T., Groot M., Howard J. C. (1997) Annu. Rev. Immunol. 15, 749–795 - PubMed
-
- Dunn G. P., Koebel C. M., Schreiber R. D. (2006) Nat. Rev. Immunol. 6, 836–848 - PubMed
-
- Schroder K., Hertzog P. J., Ravasi T., Hume D. A. (2004) J. Leukocyte Biol. 75, 163–189 - PubMed
-
- Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

