Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients
- PMID: 20682973
- PMCID: PMC3568974
Aberrant promoter hypermethylation and genomic hypomethylation in tumor, adjacent normal tissues and blood from breast cancer patients
Abstract
Background: Promoter hypermethylation and global hypomethylation in the human genome are hallmarks of most cancers. Detection of aberrant methylation in white blood cells (WBC) has been suggested as a marker for cancer development, but has not been extensively investigated. This study was carried out to determine whether aberrant methylation in WBC DNA can be used as a surrogate biomarker for breast cancer risk.
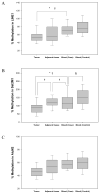
Patients and methods: Promoter hypermethylation of 8 tumor suppressor genes (RASSF1A, APC, HIN1, BRCA1, CYCLIND2, RARbeta, CDH1 and TWIST1) and DNA methylation for three repetitive elements (LINE1, Sat2 and Alu) were analyzed in invasive ductal carcinoma of the breast, paired adjacent normal tissue and WBC from 40 breast cancer patients by the MethyLight assay. Methylation in WBC from 40 controls was also analyzed.
Results: Tumor and adjacent tissues showed frequent hypermethylation for all genes tested, while WBC DNA was rarely hypermethylated. For HIN1, RASSF1A, APC and TWIST1, there was agreement between hypermethylation in tumor and adjacent tissues (p=0.04, p=0.02, p=0.005 and p<0.0001, respectively). DNA methylation for the three repetitive elements was lower in tumor compared to adjacent tissue and WBC DNA. Significant correlations in the methylation of Sat2M1 between tumor and adjacent tissues and WBC DNA were found (p<0.0001 and p=0.046, respectively). There was also a significant difference in methylation of Sat2M1 between cases and controls (p=0.01).
Conclusion: These results suggest that further studies of WBC methylation, including prospective studies, may provide biomarkers of breast cancer risk.
Figures

Similar articles
-
Blood-based DNA methylation as biomarker for breast cancer: a systematic review.Clin Epigenetics. 2016 Nov 14;8:115. doi: 10.1186/s13148-016-0282-6. eCollection 2016. Clin Epigenetics. 2016. PMID: 27895805 Free PMC article. Review.
-
[Detection of free tumor-related DNA in the serum of breast cancer patients].Zhonghua Zhong Liu Za Zhi. 2007 Aug;29(8):609-13. Zhonghua Zhong Liu Za Zhi. 2007. PMID: 18210882 Chinese.
-
Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients.Clin Cancer Res. 2004 Sep 15;10(18 Pt 1):6189-93. doi: 10.1158/1078-0432.CCR-04-0597. Clin Cancer Res. 2004. PMID: 15448006
-
Detection of promoter methylation of tumor suppressor genes in serum DNA of breast cancer cases and benign breast disease controls.Epigenetics. 2012 Nov;7(11):1258-67. doi: 10.4161/epi.22220. Epub 2012 Sep 17. Epigenetics. 2012. PMID: 22986510 Free PMC article.
-
DNA hypomethylation is prevalent even in low-grade breast cancers.Cancer Biol Ther. 2004 Dec;3(12):1225-31. doi: 10.4161/cbt.3.12.1222. Epub 2004 Dec 9. Cancer Biol Ther. 2004. PMID: 15539937 Review.
Cited by
-
DNA methylation in peripheral blood: a potential biomarker for cancer molecular epidemiology.J Epidemiol. 2012;22(5):384-94. doi: 10.2188/jea.je20120003. Epub 2012 Aug 4. J Epidemiol. 2012. PMID: 22863985 Free PMC article. Review.
-
Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort.Cancer Epidemiol Biomarkers Prev. 2011 Dec;20(12):2518-23. doi: 10.1158/1055-9965.EPI-11-0553. Epub 2011 Oct 12. Cancer Epidemiol Biomarkers Prev. 2011. PMID: 21994404 Free PMC article.
-
Genomic DNA Hypomethylation and Risk of Renal Cell Carcinoma: A Case-Control Study.Clin Cancer Res. 2016 Apr 15;22(8):2074-82. doi: 10.1158/1078-0432.CCR-15-0977. Epub 2015 Dec 11. Clin Cancer Res. 2016. PMID: 26655847 Free PMC article.
-
Blood-based DNA methylation as biomarker for breast cancer: a systematic review.Clin Epigenetics. 2016 Nov 14;8:115. doi: 10.1186/s13148-016-0282-6. eCollection 2016. Clin Epigenetics. 2016. PMID: 27895805 Free PMC article. Review.
-
HERV-K and LINE-1 DNA Methylation and Reexpression in Urothelial Carcinoma.Front Oncol. 2013 Sep 26;3:255. doi: 10.3389/fonc.2013.00255. eCollection 2013. Front Oncol. 2013. PMID: 24133654 Free PMC article.
References
-
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer Statistics, 2009. CA Cancer J Clin. 2009;59:225–249. - PubMed
-
- Rosenberg K. Ten-year risk of false positive screening mammograms and clinical breast examinations. J Nurse-Midwifery. 1998;43:394–395. - PubMed
-
- Müller HM, Widschwendter A, Fiegl H, Ivarsson L, Goebel G, Perkmann E, Marth C, Widschwendter M. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–7645. - PubMed
-
- Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. - PubMed
-
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3:253–266. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
