Free energy calculations reveal rotating-ratchet mechanism for DNA supercoil relaxation by topoisomerase IB and its inhibition
- PMID: 20682265
- PMCID: PMC2913206
- DOI: 10.1016/j.bpj.2010.04.077
Free energy calculations reveal rotating-ratchet mechanism for DNA supercoil relaxation by topoisomerase IB and its inhibition
Abstract
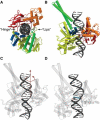
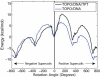
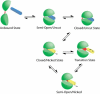
Topoisomerases maintain the proper topological state of DNA. Human topoisomerase I removes DNA supercoils by clamping a duplex DNA segment, nicking one strand at a phosphodiester bond, covalently attaching to the 3' end of the nick, and allowing the DNA downstream of the cut to rotate around the intact strand. Using molecular dynamics simulations and umbrella sampling free energy calculations, we show that the rotation of downstream DNA in the grip of the enzyme that brings about release of positive or negative supercoils occurs by thermally assisted diffusion on ratchet energy profiles. The ratchetlike free-energy-versus-rotation profile that we compute provides a model for the function of topoisomerase in which the periodic maxima along the profile modulate the rate of supercoil relaxation, while the minima provide metastable conformational states for DNA religation. The results confirm previous experimental and computational work, and suggest that relaxation of the two types of supercoils involves distinct protein pathways. Additionally, simulations performed with the ternary complex of topoisomerase, DNA, and the chemotherapeutic drug topotecan show important differences in the mechanisms for supercoil relaxation when the drug is present, accounting for the relative values of relaxation rates measured in single-molecule experiments. Good agreement is found between rate constants from tweezer experiments and those calculated from simulations. Evidence is presented for the existence of semiopen states of the protein, which facilitate rotations after the initial one, as a result of biasing the protein into a conformation more favorable to strand rotation than the closed state required for nicking of the DNA.
2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
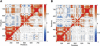
Similar articles
-
Rotation of DNA around intact strand in human topoisomerase I implies distinct mechanisms for positive and negative supercoil relaxation.Nucleic Acids Res. 2005 Nov 27;33(20):6621-34. doi: 10.1093/nar/gki935. Print 2005. Nucleic Acids Res. 2005. PMID: 16314322 Free PMC article.
-
Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB.Nature. 2005 Mar 31;434(7033):671-4. doi: 10.1038/nature03395. Nature. 2005. PMID: 15800630
-
Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation.Biochemistry. 1997 Apr 29;36(17):5212-22. doi: 10.1021/bi962880t. Biochemistry. 1997. PMID: 9136883
-
Cellular strategies for regulating DNA supercoiling: a single-molecule perspective.Cell. 2010 Aug 20;142(4):519-30. doi: 10.1016/j.cell.2010.08.001. Cell. 2010. PMID: 20723754 Free PMC article. Review.
-
Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs.Curr Med Chem. 2010;17(35):4270-90. doi: 10.2174/092986710793361252. Curr Med Chem. 2010. PMID: 20939813 Review.
Cited by
-
Distant residues modulate conformational opening in SARS-CoV-2 spike protein.Proc Natl Acad Sci U S A. 2021 Oct 26;118(43):e2100943118. doi: 10.1073/pnas.2100943118. Epub 2021 Oct 6. Proc Natl Acad Sci U S A. 2021. PMID: 34615730 Free PMC article.
-
Simulation of DNA Supercoil Relaxation.Biophys J. 2016 May 24;110(10):2176-84. doi: 10.1016/j.bpj.2016.03.041. Biophys J. 2016. PMID: 27224483 Free PMC article.
-
The dynamic interplay between DNA topoisomerases and DNA topology.Biophys Rev. 2016 Nov;8(Suppl 1):101-111. doi: 10.1007/s12551-016-0240-8. Epub 2016 Nov 14. Biophys Rev. 2016. PMID: 28510219 Free PMC article. Review.
-
Slowdown of Interhelical Motions Induces a Glass Transition in RNA.Biophys J. 2015 Jun 16;108(12):2876-85. doi: 10.1016/j.bpj.2015.04.041. Biophys J. 2015. PMID: 26083927 Free PMC article.
-
A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity.Proc Natl Acad Sci U S A. 2012 Oct 2;109(40):16125-30. doi: 10.1073/pnas.1206480109. Epub 2012 Sep 18. Proc Natl Acad Sci U S A. 2012. PMID: 22991469 Free PMC article.
References
-
- Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. - PubMed
-
- Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. - PubMed
-
- Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. - PubMed
-
- Schoeffler A.J., Berger J.M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. - PubMed
-
- Leppard J.B., Champoux J.J. Human DNA topoisomerase I: relaxation, roles, and damage control. Chromosoma. 2005;114:75–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

