Hrs controls sorting of the epithelial Na+ channel between endosomal degradation and recycling pathways
- PMID: 20675381
- PMCID: PMC2945546
- DOI: 10.1074/jbc.M110.150755
Hrs controls sorting of the epithelial Na+ channel between endosomal degradation and recycling pathways
Abstract
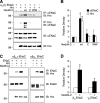
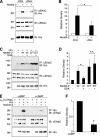
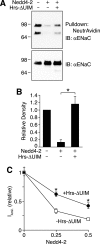
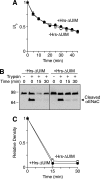
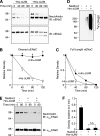
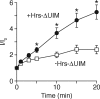
Epithelial Na(+) absorption is regulated by Nedd4-2, an E3 ubiquitin ligase that reduces expression of the epithelial Na(+) channel (ENaC) at the cell surface. Defects in this regulation cause Liddle syndrome, an inherited form of hypertension. Previous work found that Nedd4-2 functions through two distinct effects on trafficking, enhancing both ENaC endocytosis and ENaC degradation in lysosomes. To investigate the mechanism by which Nedd4-2 targets ENaC to lysosomes, we tested the role of hepatocyte growth factor-regulated tyrosine kinase substrate (Hrs), a component of the endosomal sorting complexes required for transport (ESCRT)-0 complex. We found that α-, β-, and γENaC each interact with Hrs. These interactions were enhanced by Nedd4-2 and were dependent on the catalytic function of Nedd4-2 as well as its WW domains. Mutation of ENaC PY motifs, responsible for inherited hypertension (Liddle syndrome), decreased Hrs binding to ENaC. Moreover, binding of ENaC to Hrs was reduced by dexamethasone/serum- and glucocorticoid-inducible kinase and cAMP, which are signaling pathways that inhibit Nedd4-2. Nedd4-2 bound to Hrs and catalyzed Hrs ubiquitination but did not alter Hrs protein levels. Expression of a dominant negative Hrs lacking its ubiquitin-interacting motif (Hrs-ΔUIM) increased ENaC surface expression and current. This occurred through reduced degradation of the cell surface pool of proteolytically activated ENaC, which enhanced its recycling to the cell surface. In contrast, Hrs-ΔUIM had no effect on degradation of uncleaved inactive channels. The data support a model in which Nedd4-2 induces binding of ENaC to Hrs, which mediates the sorting decision between ENaC degradation and recycling.
Figures
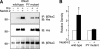
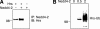
Similar articles
-
Nedd4-2 catalyzes ubiquitination and degradation of cell surface ENaC.J Biol Chem. 2007 Jul 13;282(28):20207-12. doi: 10.1074/jbc.M611329200. Epub 2007 May 14. J Biol Chem. 2007. PMID: 17502380
-
Nedd4-2 induces endocytosis and degradation of proteolytically cleaved epithelial Na+ channels.J Biol Chem. 2008 Mar 7;283(10):6033-9. doi: 10.1074/jbc.M708555200. Epub 2008 Jan 3. J Biol Chem. 2008. PMID: 18174164
-
The PY motif of ENaC, mutated in Liddle syndrome, regulates channel internalization, sorting and mobilization from subapical pool.Traffic. 2007 Sep;8(9):1246-64. doi: 10.1111/j.1600-0854.2007.00602.x. Epub 2007 Jul 1. Traffic. 2007. PMID: 17605762
-
Role of the UPS in Liddle syndrome.BMC Biochem. 2008 Oct 21;9 Suppl 1(Suppl 1):S5. doi: 10.1186/1471-2091-9-S1-S5. BMC Biochem. 2008. PMID: 19007435 Free PMC article. Review.
-
Regulation of the epithelial Na+ channel by Nedd4 and ubiquitination.Kidney Int. 2000 Mar;57(3):809-15. doi: 10.1046/j.1523-1755.2000.00919.x. Kidney Int. 2000. PMID: 10720933 Review.
Cited by
-
Ubiquitin-specific peptidase 8 (USP8) regulates endosomal trafficking of the epithelial Na+ channel.J Biol Chem. 2013 Feb 22;288(8):5389-97. doi: 10.1074/jbc.M112.425272. Epub 2013 Jan 7. J Biol Chem. 2013. PMID: 23297398 Free PMC article.
-
Acetylation stimulates the epithelial sodium channel by reducing its ubiquitination and degradation.J Biol Chem. 2015 May 15;290(20):12497-503. doi: 10.1074/jbc.M114.635540. Epub 2015 Mar 18. J Biol Chem. 2015. PMID: 25787079 Free PMC article.
-
The epithelial sodium channel (ENaC) establishes a trafficking vesicle pool responsible for its regulation.PLoS One. 2012;7(9):e46593. doi: 10.1371/journal.pone.0046593. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029554 Free PMC article.
-
ENaC structure and function in the wake of a resolved structure of a family member.Am J Physiol Renal Physiol. 2011 Oct;301(4):F684-96. doi: 10.1152/ajprenal.00259.2011. Epub 2011 Jul 13. Am J Physiol Renal Physiol. 2011. PMID: 21753073 Free PMC article. Review.
-
SMURF and NEDD4: sharp shooters monitor the gate keepers and ion traffic controllers of lead astray cell.J Membr Biol. 2011 Nov;244(1):1-8. doi: 10.1007/s00232-011-9394-2. Epub 2011 Sep 15. J Membr Biol. 2011. PMID: 21918841 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

