The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response
- PMID: 20670894
- PMCID: PMC2928996
- DOI: 10.1016/j.molcel.2010.07.005
The FANCM/FAAP24 complex is required for the DNA interstrand crosslink-induced checkpoint response
Abstract
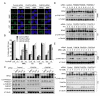
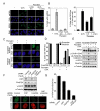
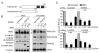
Cells from Fanconi anemia (FA) patients are extremely sensitive to DNA interstrand crosslinking (ICL) agents, but the molecular basis of the hypersensitivity remains to be explored. FANCM (FA complementation group M), and its binding partner, FAAP24, anchor the multisubunit FA core complex to chromatin after DNA damage and may contribute to ICL-specific cellular response. Here we show that the FANCM/FAAP24 complex is specifically required for the recruitment of replication protein A (RPA) to ICL-stalled replication forks. ICL-induced RPA foci formation requires the DNA-binding activity of FAAP24 but not the DNA translocase activity of FANCM. Furthermore, FANCM/FAAP24-dependent RPA foci formation is required for efficient ATR-mediated checkpoint activation in response to ICL. Therefore, we propose that FANCM/FAAP24 plays a role in ICL-induced checkpoint activation through regulating RPA recruiment at ICL-stalled replication forks.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
And-1 Coordinates with the FANCM Complex to Regulate Fanconi Anemia Signaling and Cisplatin Resistance.Cancer Res. 2022 Sep 16;82(18):3249-3262. doi: 10.1158/0008-5472.CAN-22-0769. Cancer Res. 2022. PMID: 35867033 Free PMC article.
-
FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex.Mol Cell. 2008 Nov 7;32(3):313-24. doi: 10.1016/j.molcel.2008.10.014. Mol Cell. 2008. PMID: 18995830
-
FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions.Mol Cell. 2013 Mar 7;49(5):997-1009. doi: 10.1016/j.molcel.2012.12.010. Epub 2013 Jan 17. Mol Cell. 2013. PMID: 23333308 Free PMC article.
-
FANCM-FAAP24 and HCLK2: roles in ATR signalling and the Fanconi anemia pathway.Cell Cycle. 2009 Apr 15;8(8):1133-7. doi: 10.4161/cc.8.8.8204. Epub 2009 Apr 16. Cell Cycle. 2009. PMID: 19282663 Review.
-
FANCM-FAAP24 and FANCJ: FA proteins that metabolize DNA.Mutat Res. 2009 Jul 31;668(1-2):20-6. doi: 10.1016/j.mrfmmm.2009.04.002. Epub 2009 Apr 18. Mutat Res. 2009. PMID: 19379763 Free PMC article. Review.
Cited by
-
Biochemical Activities and Genetic Functions of the Drosophila melanogaster Fancm Helicase in DNA Repair.Genetics. 2016 Oct;204(2):531-541. doi: 10.1534/genetics.116.192534. Epub 2016 Jul 27. Genetics. 2016. PMID: 27466228 Free PMC article.
-
The Adaptive Mechanisms and Checkpoint Responses to a Stressed DNA Replication Fork.Int J Mol Sci. 2023 Jun 22;24(13):10488. doi: 10.3390/ijms241310488. Int J Mol Sci. 2023. PMID: 37445667 Free PMC article. Review.
-
The MCM8/9 complex: A recent recruit to the roster of helicases involved in genome maintenance.DNA Repair (Amst). 2019 Apr;76:1-10. doi: 10.1016/j.dnarep.2019.02.003. Epub 2019 Feb 5. DNA Repair (Amst). 2019. PMID: 30743181 Free PMC article. Review.
-
Stress and DNA repair biology of the Fanconi anemia pathway.Blood. 2014 Oct 30;124(18):2812-9. doi: 10.1182/blood-2014-04-526293. Epub 2014 Sep 18. Blood. 2014. PMID: 25237197 Free PMC article. Review.
-
Modularized functions of the Fanconi anemia core complex.Cell Rep. 2014 Jun 26;7(6):1849-57. doi: 10.1016/j.celrep.2014.04.029. Epub 2014 Jun 5. Cell Rep. 2014. PMID: 24910428 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

