Characterization of the envelope glycoproteins associated with infectious hepatitis C virus
- PMID: 20668082
- PMCID: PMC2937754
- DOI: 10.1128/JVI.01180-10
Characterization of the envelope glycoproteins associated with infectious hepatitis C virus
Abstract
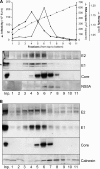
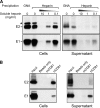
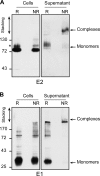
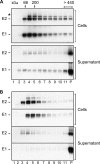
Hepatitis C is caused by an enveloped virus whose entry is mediated by two glycoproteins, namely, E1 and E2, which have been shown to assemble as a noncovalent heterodimer. Despite extensive research in the field of such an important human pathogen, hepatitis C virus (HCV) glycoproteins have only been studied so far in heterologous expression systems, and their organization at the surfaces of infectious virions has not yet been described. Here, we characterized the envelope glycoproteins associated with cell-cultured infectious virions and compared them with their prebudding counterparts. Viral particles were analyzed by ultracentrifugation, and the envelope glycoproteins were characterized by coimmunoprecipitation and receptor pulldown assays. Furthermore, their oligomeric state was determined by sedimentation through sucrose gradients and by separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. In sucrose gradient analyses, HCV envelope glycoproteins were associated with fractions containing the most infectious viral particles. Importantly, besides maturation of some of their glycans, HCV envelope glycoproteins showed a dramatic change in their oligomeric state after incorporation into the viral particle. Indeed, virion-associated E1 and E2 envelope glycoproteins formed large covalent complexes stabilized by disulfide bridges, whereas the intracellular forms of these proteins assembled as noncovalent heterodimers. Furthermore, the virion-associated glycoprotein complexes were recognized by the large extracellular loop of CD81 as well as conformation-sensitive antibodies, indicating that these proteins are in a functional conformation. Overall, our study fills a gap in the description of HCV outer morphology and should guide further investigations into virus entry and assembly.
Figures



Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
Cited by
-
Interplay between basic residues of hepatitis C virus glycoprotein E2 with viral receptors, neutralizing antibodies and lipoproteins.PLoS One. 2012;7(12):e52651. doi: 10.1371/journal.pone.0052651. Epub 2012 Dec 27. PLoS One. 2012. PMID: 23300734 Free PMC article.
-
Immunoregulation through membrane proteins modified by reducing conditions induced by immune reactions.Eur J Immunol. 2013 Jan;43(1):15-21. doi: 10.1002/eji.201242849. Eur J Immunol. 2013. PMID: 23233323 Free PMC article. Review.
-
Role of conserved cysteine residues in hepatitis C virus glycoprotein e2 folding and function.J Virol. 2012 Apr;86(7):3961-74. doi: 10.1128/JVI.05396-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278231 Free PMC article.
-
Hepatitis C virus assembly imaging.Viruses. 2011 Nov;3(11):2238-54. doi: 10.3390/v3112238. Epub 2011 Nov 15. Viruses. 2011. PMID: 22163343 Free PMC article. Review.
-
HCV glycoprotein structures: what to expect from the unexpected.Curr Opin Virol. 2015 Jun;12:53-8. doi: 10.1016/j.coviro.2015.02.004. Epub 2015 Mar 16. Curr Opin Virol. 2015. PMID: 25790756 Free PMC article. Review.
References
-
- Bartenschlager, R. 2005. The hepatitis C virus replicon system: from basic research to clinical application. J. Hepatol. 43:210-216. - PubMed
-
- Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 278:41003-41012. - PubMed
-
- Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. U. S. A. 100:14199-14204. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

