The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis
- PMID: 20667826
- PMCID: PMC2975206
- DOI: 10.1074/jbc.M110.131623
The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis
Abstract
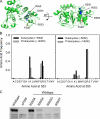
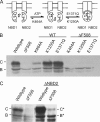
The deletion of phenylalanine 508 in the first nucleotide binding domain of the cystic fibrosis transmembrane conductance regulator is directly associated with >90% of cystic fibrosis cases. This mutant protein fails to traffic out of the endoplasmic reticulum and is subsequently degraded by the proteasome. The effects of this mutation may be partially reversed by the application of exogenous osmolytes, expression at low temperature, and the introduction of second site suppressor mutations. However, the specific steps of folding and assembly of full-length cystic fibrosis transmembrane conductance regulator (CFTR) directly altered by the disease-causing mutation are unclear. To elucidate the effects of the ΔF508 mutation, on various steps in CFTR folding, a series of misfolding and suppressor mutations in the nucleotide binding and transmembrane domains were evaluated for effects on the folding and maturation of the protein. The results indicate that the isolated NBD1 responds to both the ΔF508 mutation and intradomain suppressors of this mutation. In addition, identification of a novel second site suppressor of the defect within the second transmembrane domain suggests that ΔF508 also effects interdomain interactions critical for later steps in the biosynthesis of CFTR.
Figures
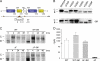


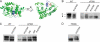
Similar articles
-
Impact of the deltaF508 mutation in first nucleotide-binding domain of human cystic fibrosis transmembrane conductance regulator on domain folding and structure.J Biol Chem. 2005 Jan 14;280(2):1346-53. doi: 10.1074/jbc.M410968200. Epub 2004 Nov 3. J Biol Chem. 2005. PMID: 15528182
-
The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR.Nat Struct Mol Biol. 2005 Jan;12(1):17-25. doi: 10.1038/nsmb882. Epub 2004 Dec 26. Nat Struct Mol Biol. 2005. PMID: 15619635
-
Restoration of NBD1 thermal stability is necessary and sufficient to correct ∆F508 CFTR folding and assembly.J Mol Biol. 2015 Jan 16;427(1):106-20. doi: 10.1016/j.jmb.2014.07.026. Epub 2014 Jul 30. J Mol Biol. 2015. PMID: 25083918 Free PMC article.
-
CFTR: folding, misfolding and correcting the ΔF508 conformational defect.Trends Mol Med. 2012 Feb;18(2):81-91. doi: 10.1016/j.molmed.2011.10.003. Epub 2011 Dec 3. Trends Mol Med. 2012. PMID: 22138491 Free PMC article. Review.
-
Control of cystic fibrosis transmembrane conductance regulator membrane trafficking: not just from the endoplasmic reticulum to the Golgi.FEBS J. 2013 Sep;280(18):4396-406. doi: 10.1111/febs.12392. Epub 2013 Jul 5. FEBS J. 2013. PMID: 23773658 Review.
Cited by
-
Cystic fibrosis transmembrane conductance regulator (ABCC7) structure.Cold Spring Harb Perspect Med. 2013 Feb 1;3(2):a009514. doi: 10.1101/cshperspect.a009514. Cold Spring Harb Perspect Med. 2013. PMID: 23378596 Free PMC article. Review.
-
Cystic fibrosis transmembrane regulator correctors and potentiators.Cold Spring Harb Perspect Med. 2013 Jul 1;3(7):a009761. doi: 10.1101/cshperspect.a009761. Cold Spring Harb Perspect Med. 2013. PMID: 23818513 Free PMC article. Review.
-
Understanding CFTR Functionality: A Comprehensive Review of Tests and Modulator Therapy in Cystic Fibrosis.Cell Biochem Biophys. 2024 Mar;82(1):15-34. doi: 10.1007/s12013-023-01200-w. Epub 2023 Dec 4. Cell Biochem Biophys. 2024. PMID: 38048024 Review.
-
Structural Comparative Modeling of Multi-Domain F508del CFTR.Biomolecules. 2022 Mar 18;12(3):471. doi: 10.3390/biom12030471. Biomolecules. 2022. PMID: 35327663 Free PMC article.
-
Alteration of protein function by a silent polymorphism linked to tRNA abundance.PLoS Biol. 2017 May 16;15(5):e2000779. doi: 10.1371/journal.pbio.2000779. eCollection 2017 May. PLoS Biol. 2017. PMID: 28510592 Free PMC article.
References
-
- Minetti C. A., Remeta D. P. (2006) Arch. Biochem. Biophys. 453, 32–53 - PubMed
-
- Jansens A., van Duijn E., Braakman I. (2002) Science 298, 2401–2403 - PubMed
-
- Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. (1989) Science 245, 1066–1073 - PubMed
-
- Lewis H. A., Buchanan S. G., Burley S. K., Conners K., Dickey M., Dorwart M., Fowler R., Gao X., Guggino W. B., Hendrickson W. A., Hunt J. F., Kearins M. C., Lorimer D., Maloney P. C., Post K. W., Rajashankar K. R., Rutter M. E., Sauder J. M., Shriver S., Thibodeau P. H., Thomas P. J., Zhang M., Zhao X., Emtage S. (2004) EMBO J. 23, 282–293 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

