Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism
- PMID: 20660762
- PMCID: PMC2941284
- DOI: 10.1073/pnas.1002176107
Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism
Abstract
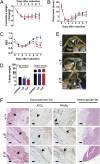
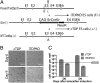
Tat activating regulatory DNA-binding protein (Tardbp or TDP-43), a highly conserved metazoan DNA/RNA binding protein thought to be involved in RNA transcription and splicing, has been linked to the pathophysiology of amyotrophic lateral sclerosis and frontotemporal lobar degeneration and is essential for early embryonic development. However, neither the physiological role of TDP-43 in the adult nor its downstream targets are well defined. To address these questions, we developed conditional Tardbp-KO mice and embryonic stem (ES) cell models. Here, we show that postnatal deletion of Tardbp in mice caused dramatic loss of body fat followed by rapid death. Moreover, conditional Tardbp-KO ES cells failed to proliferate. Importantly, high-throughput DNA sequencing analysis on the transcriptome of ES cells lacking Tardbp revealed a set of downstream targets of TDP-43. We show that Tbc1d1, a gene known to mediate leanness and linked to obesity, is down-regulated in the absence of TDP-43. Collectively, our results establish that TDP-43 is critical for fat metabolism and ES cell survival.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
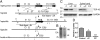

Similar articles
-
TDP-43, an ALS linked protein, regulates fat deposition and glucose homeostasis.PLoS One. 2013 Aug 13;8(8):e71793. doi: 10.1371/journal.pone.0071793. eCollection 2013. PLoS One. 2013. PMID: 23967244 Free PMC article.
-
TDP-43 is a developmentally regulated protein essential for early embryonic development.J Biol Chem. 2010 Feb 26;285(9):6826-34. doi: 10.1074/jbc.M109.061846. Epub 2009 Dec 29. J Biol Chem. 2010. PMID: 20040602 Free PMC article.
-
Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis.Acta Neuropathol. 2010 Apr;119(4):409-19. doi: 10.1007/s00401-010-0659-0. Epub 2010 Mar 3. Acta Neuropathol. 2010. PMID: 20198480 Free PMC article.
-
Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: disease-associated pathways.Rev Neurosci. 2010;21(4):251-72. doi: 10.1515/revneuro.2010.21.4.251. Rev Neurosci. 2010. PMID: 21086759 Review.
-
Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins.J Neurochem. 2016 Aug;138 Suppl 1:95-111. doi: 10.1111/jnc.13625. Epub 2016 Jun 15. J Neurochem. 2016. PMID: 27015757 Review.
Cited by
-
AMPK Signalling and Defective Energy Metabolism in Amyotrophic Lateral Sclerosis.Neurochem Res. 2016 Mar;41(3):544-53. doi: 10.1007/s11064-015-1665-3. Epub 2015 Jul 23. Neurochem Res. 2016. PMID: 26202426 Review.
-
FUS Mutation Causes Disordered Lipid Metabolism in Skeletal Muscle Associated with ALS.Mol Neurobiol. 2022 Dec;59(12):7265-7277. doi: 10.1007/s12035-022-03048-2. Epub 2022 Sep 28. Mol Neurobiol. 2022. PMID: 36169888
-
Status of ALS Treatment, Insights into Therapeutic Challenges and Dilemmas.J Pers Med. 2022 Sep 28;12(10):1601. doi: 10.3390/jpm12101601. J Pers Med. 2022. PMID: 36294741 Free PMC article. Review.
-
TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD.Science. 2015 Aug 7;349(6248):650-5. doi: 10.1126/science.aab0983. Science. 2015. PMID: 26250685 Free PMC article.
-
TDP-43/FUS in motor neuron disease: Complexity and challenges.Prog Neurobiol. 2016 Oct-Nov;145-146:78-97. doi: 10.1016/j.pneurobio.2016.09.004. Epub 2016 Sep 28. Prog Neurobiol. 2016. PMID: 27693252 Free PMC article. Review.
References
-
- Buratti E, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: An important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. - PubMed
-
- Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

