Three-dimensional structure of Rubella virus factories
- PMID: 20655079
- PMCID: PMC7111912
- DOI: 10.1016/j.virol.2010.06.043
Three-dimensional structure of Rubella virus factories
Abstract
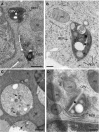
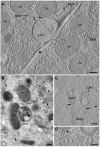
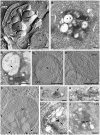
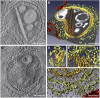
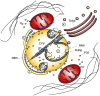
Viral factories are complex structures in the infected cell where viruses compartmentalize their life cycle. Rubella virus (RUBV) assembles factories by recruitment of rough endoplasmic reticulum (RER), mitochondria and Golgi around modified lysosomes known as cytopathic vacuoles or CPVs. These organelles contain active replication complexes that transfer replicated RNA to assembly sites in Golgi membranes. We have studied the structure of RUBV factory in three dimensions by electron tomography and freeze-fracture. CPVs contain stacked membranes, rigid sheets, small vesicles and large vacuoles. These membranes are interconnected and in communication with the endocytic pathway since they incorporate endocytosed BSA-gold. RER and CPVs are coupled through protein bridges and closely apposed membranes. Golgi vesicles attach to the CPVs but no tight contacts with mitochondria were detected. Immunogold labelling confirmed that the mitochondrial protein p32 is an abundant component around and inside CPVs where it could play important roles in factory activities.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Novel replication complex architecture in rubella replicon-transfected cells.Cell Microbiol. 2007 Apr;9(4):875-90. doi: 10.1111/j.1462-5822.2006.00837.x. Epub 2006 Nov 3. Cell Microbiol. 2007. PMID: 17087733 Free PMC article.
-
Structural maturation of rubella virus in the Golgi complex.Virology. 2003 Aug 1;312(2):261-9. doi: 10.1016/s0042-6822(03)00384-2. Virology. 2003. PMID: 12919732 Free PMC article.
-
The rubella virus E1 glycoprotein is arrested in a novel post-ER, pre-Golgi compartment.J Cell Biol. 1992 Aug;118(4):795-811. doi: 10.1083/jcb.118.4.795. J Cell Biol. 1992. PMID: 1500424 Free PMC article.
-
Biogenesis and architecture of arterivirus replication organelles.Virus Res. 2016 Jul 15;220:70-90. doi: 10.1016/j.virusres.2016.04.001. Epub 2016 Apr 9. Virus Res. 2016. PMID: 27071852 Free PMC article. Review.
-
Endoplasmic Reticulum: The Favorite Intracellular Niche for Viral Replication and Assembly.Viruses. 2016 Jun 7;8(6):160. doi: 10.3390/v8060160. Viruses. 2016. PMID: 27338443 Free PMC article. Review.
Cited by
-
Ultrastructure of the replication sites of positive-strand RNA viruses.Virology. 2015 May;479-480:418-33. doi: 10.1016/j.virol.2015.02.029. Epub 2015 Mar 6. Virology. 2015. PMID: 25746936 Free PMC article. Review.
-
Role of host reticulon proteins in rearranging membranes for positive-strand RNA virus replication.Curr Opin Microbiol. 2012 Aug;15(4):519-24. doi: 10.1016/j.mib.2012.04.007. Epub 2012 May 21. Curr Opin Microbiol. 2012. PMID: 22621853 Free PMC article. Review.
-
Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes.mBio. 2014 Feb 18;5(1):e00931-13. doi: 10.1128/mBio.00931-13. mBio. 2014. PMID: 24549844 Free PMC article.
-
Viral infection: Moving through complex and dynamic cell-membrane structures.Commun Integr Biol. 2011 Jul;4(4):398-408. doi: 10.4161/cib.4.4.16716. Epub 2011 Jul 1. Commun Integr Biol. 2011. PMID: 21966556 Free PMC article.
-
The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments.mBio. 2011 Oct 4;2(5):e00166-11. doi: 10.1128/mBio.00166-11. Print 2011. mBio. 2011. PMID: 21972238 Free PMC article.
References
-
- Cabezas P., Risco C. Studying cellular architecture in three dimensions with improved resolution: Ta replicas revisited. Cell Biol. Int. 2006;30:747–754. - PubMed
-
- Cardone G., Grünewald K., Steven A.C. A resolution criterion for electron tomography based on cross-validation. J. Struct. Biol. 2005;151:117–129. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

