A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography
- PMID: 20639890
- PMCID: PMC2917476
- DOI: 10.1038/nm.2185
A molecularly engineered split reporter for imaging protein-protein interactions with positron emission tomography
Abstract
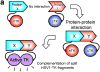
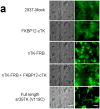
Improved techniques to noninvasively image protein-protein interactions (PPIs) are essential. We molecularly engineered a positron emission tomography (PET)-based split reporter (herpes simplex virus type 1 thymidine kinase), cleaved between Thr265 and Ala266, and used this in a protein-fragment complementation assay (PCA) to quantify PPIs in mammalian cells and to microPET image them in living mice. An introduced point mutation (V119C) markedly enhanced thymidine kinase complementation in PCAs, on the basis of rapamycin modulation of FKBP12-rapamycin-binding domain (FRB) and FKBP12 (FK506 binding protein), the interaction of hypoxia-inducible factor-1alpha with the von Hippel-Lindau tumor suppressor, and in an estrogen receptor intramolecular protein folding assay. Applications of this unique split thymidine kinase are potentially far reaching, including, for example, considerably more accurate monitoring of immune and stem cell therapies, allowing for fully quantitative and tomographic PET localization of PPIs in preclinical small- and large-animal models of disease.
Figures









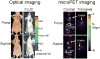
Similar articles
-
Structure-guided engineering of human thymidine kinase 2 as a positron emission tomography reporter gene for enhanced phosphorylation of non-natural thymidine analog reporter probe.J Biol Chem. 2012 Jan 2;287(1):446-454. doi: 10.1074/jbc.M111.314666. Epub 2011 Nov 9. J Biol Chem. 2012. PMID: 22074768 Free PMC article. Clinical Trial.
-
Thymidine Kinase PET Reporter Gene Imaging of Cancer Cells In Vivo.Methods Mol Biol. 2018;1790:137-151. doi: 10.1007/978-1-4939-7860-1_11. Methods Mol Biol. 2018. PMID: 29858789
-
Molecular imaging of drug-modulated protein-protein interactions in living subjects.Cancer Res. 2004 Mar 15;64(6):2113-9. doi: 10.1158/0008-5472.can-03-2972. Cancer Res. 2004. PMID: 15026351 Free PMC article.
-
Beyond the hypoxia-inducible factor-centric tumour suppressor model of von Hippel-Lindau.Curr Opin Oncol. 2008 Jan;20(1):83-9. doi: 10.1097/CCO.0b013e3282f310de. Curr Opin Oncol. 2008. PMID: 18043261 Review.
-
Protein-fragment complementation assays for large-scale analysis of protein-protein interactions.Biochem Soc Trans. 2021 Jun 30;49(3):1337-1348. doi: 10.1042/BST20201058. Biochem Soc Trans. 2021. PMID: 34156434 Free PMC article. Review.
Cited by
-
Quantification and tracking of genetically engineered dendritic cells for studying immunotherapy.Magn Reson Med. 2018 Feb;79(2):1010-1019. doi: 10.1002/mrm.26708. Epub 2017 May 7. Magn Reson Med. 2018. PMID: 28480589 Free PMC article.
-
Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system.Microbiol Mol Biol Rev. 2012 Jun;76(2):331-82. doi: 10.1128/MMBR.05021-11. Microbiol Mol Biol Rev. 2012. PMID: 22688816 Free PMC article. Review.
-
Live-cell imaging of single mRNA dynamics using split superfolder green fluorescent proteins with minimal background.RNA. 2020 Jan;26(1):101-109. doi: 10.1261/rna.067835.118. Epub 2019 Oct 22. RNA. 2020. PMID: 31641028 Free PMC article.
-
Split-protein systems: beyond binary protein-protein interactions.Curr Opin Chem Biol. 2011 Dec;15(6):789-97. doi: 10.1016/j.cbpa.2011.10.014. Epub 2011 Nov 7. Curr Opin Chem Biol. 2011. PMID: 22070901 Free PMC article. Review.
-
Genetically encoded molecular biosensors to image histone methylation in living animals.Anal Chem. 2015 Jan 20;87(2):892-9. doi: 10.1021/ac502629r. Epub 2014 Dec 24. Anal Chem. 2015. PMID: 25506787 Free PMC article.
References
-
- Valdar WS, Thornton JM. Protein-protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001;42:108–124. - PubMed
-
- Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Devel. 2003;17:545–580. - PubMed
-
- Michnick SW. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr Opin Struct Biol. 2001;11:472–477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

