The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain
- PMID: 20637642
- PMCID: PMC2919622
- DOI: 10.1016/j.str.2010.05.007
The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain
Abstract
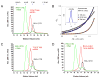
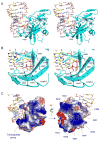
RIG-I is a cytosolic sensor of viral RNA that plays crucial roles in the induction of type I interferons. The C-terminal domain (CTD) of RIG-I is responsible for the recognition of viral RNA with 5' triphosphate (ppp). However, the mechanism of viral RNA recognition by RIG-I is still not fully understood. Here, we show that RIG-I CTD binds 5' ppp dsRNA or ssRNA, as well as blunt-ended dsRNA, and exhibits the highest affinity for 5' ppp dsRNA. Crystal structures of RIG-I CTD bound to 5' ppp dsRNA with GC- and AU-rich sequences revealed that RIG-I recognizes the termini of the dsRNA and interacts with the 5' ppp through extensive electrostatic interactions. Mutagenesis and RNA-binding studies demonstrated that similar binding surfaces are involved in the recognition of different forms of RNA. Mutations of key residues at the RNA-binding surface affected RIG-I signaling in cells.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

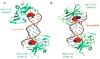
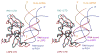
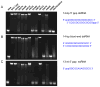
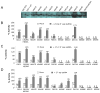
Comment in
-
RIG-I "sees" the 5'-triphosphate.Structure. 2010 Aug 11;18(8):894-6. doi: 10.1016/j.str.2010.07.002. Structure. 2010. PMID: 20696389 Free PMC article.
Similar articles
-
A structure-based model of RIG-I activation.RNA. 2012 Dec;18(12):2118-27. doi: 10.1261/rna.035949.112. Epub 2012 Nov 1. RNA. 2012. PMID: 23118418 Free PMC article. Review.
-
Structural insights into RNA recognition and activation of RIG-I-like receptors.Curr Opin Struct Biol. 2012 Jun;22(3):297-303. doi: 10.1016/j.sbi.2012.03.011. Epub 2012 May 2. Curr Opin Struct Biol. 2012. PMID: 22560447 Free PMC article. Review.
-
Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate.Nucleic Acids Res. 2011 Mar;39(4):1565-75. doi: 10.1093/nar/gkq974. Epub 2010 Oct 20. Nucleic Acids Res. 2011. PMID: 20961956 Free PMC article.
-
Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5.Arch Biochem Biophys. 2009 Aug 1;488(1):23-33. doi: 10.1016/j.abb.2009.06.008. Epub 2009 Jun 14. Arch Biochem Biophys. 2009. PMID: 19531363
-
Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors.J Biol Chem. 2009 Jun 26;284(26):17465-74. doi: 10.1074/jbc.M109.007179. Epub 2009 Apr 20. J Biol Chem. 2009. PMID: 19380577 Free PMC article.
Cited by
-
Unraveling blunt-end RNA binding and ATPase-driven translocation activities of the RIG-I family helicase LGP2.Nucleic Acids Res. 2024 Jan 11;52(1):355-369. doi: 10.1093/nar/gkad1106. Nucleic Acids Res. 2024. PMID: 38015453 Free PMC article.
-
A structure-based model of RIG-I activation.RNA. 2012 Dec;18(12):2118-27. doi: 10.1261/rna.035949.112. Epub 2012 Nov 1. RNA. 2012. PMID: 23118418 Free PMC article. Review.
-
PRRs are watching you: Localization of innate sensing and signaling regulators.Virology. 2015 May;479-480:104-9. doi: 10.1016/j.virol.2015.02.051. Epub 2015 Mar 20. Virology. 2015. PMID: 25800355 Free PMC article. Review.
-
Structural insights into RNA recognition and activation of RIG-I-like receptors.Curr Opin Struct Biol. 2012 Jun;22(3):297-303. doi: 10.1016/j.sbi.2012.03.011. Epub 2012 May 2. Curr Opin Struct Biol. 2012. PMID: 22560447 Free PMC article. Review.
-
Advancements in tyrosine kinase-mediated regulation of innate nucleic acid sensing.Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024 Jan 19;53(1):35-46. doi: 10.3724/zdxbyxb-2023-0480. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38426691 Free PMC article. Review. Chinese, English.
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. - PubMed
-
- Cui S, Eisenacher K, Kirchhofer A, Brzozka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

