beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1
- PMID: 20631169
- PMCID: PMC6632424
- DOI: 10.1523/JNEUROSCI.2154-10.2010
beta-Amyloid disrupts activity-dependent gene transcription required for memory through the CREB coactivator CRTC1
Abstract
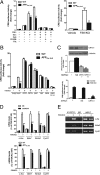
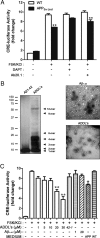
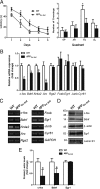
Activity-dependent gene expression mediating changes of synaptic efficacy is important for memory storage, but the mechanisms underlying gene transcriptional changes in age-related memory disorders are poorly understood. In this study, we report that gene transcription mediated by the cAMP-response element binding protein (CREB)-regulated transcription coactivator CRTC1 is impaired in neurons and brain from an Alzheimer's disease (AD) transgenic mouse expressing the human beta-amyloid precursor protein (APP(Sw,Ind)). Suppression of CRTC1-dependent gene transcription by beta-amyloid (Abeta) in response to cAMP and Ca(2+) signals is mediated by reduced calcium influx and disruption of PP2B/calcineurin-dependent CRTC1 dephosphorylation at Ser151. Consistently, expression of CRTC1 or active CRTC1 S151A and calcineurin mutants reverse the deficits on CRTC1 transcriptional activity in APP(Sw,Ind) neurons. Inhibition of calcium influx by pharmacological blockade of L-type voltage-gated calcium channels (VGCCs), but not by blocking NMDA or AMPA receptors, mimics the decrease on CRTC1 transcriptional activity observed in APP(Sw,Ind) neurons, whereas agonists of L-type VGCCs reverse efficiently these deficits. Consistent with a role of CRTC1 on Abeta-induced synaptic and memory dysfunction, we demonstrate a selective reduction of CRTC1-dependent genes related to memory (Bdnf, c-fos, and Nr4a2) coinciding with hippocampal-dependent spatial memory deficits in APP(Sw,Ind) mice. These findings suggest that CRTC1 plays a key role in coupling synaptic activity to gene transcription required for hippocampal-dependent memory, and that Abeta could disrupt cognition by affecting CRTC1 function.
Figures


Similar articles
-
CREB-regulated transcription coactivator 1-dependent transcription in Alzheimer's disease mice.Neurodegener Dis. 2012;10(1-4):250-2. doi: 10.1159/000333341. Epub 2012 Jan 6. Neurodegener Dis. 2012. PMID: 22236576
-
Crtc1 activates a transcriptional program deregulated at early Alzheimer's disease-related stages.J Neurosci. 2014 Apr 23;34(17):5776-87. doi: 10.1523/JNEUROSCI.5288-13.2014. J Neurosci. 2014. PMID: 24760838 Free PMC article.
-
CRTC1 Function During Memory Encoding Is Disrupted in Neurodegeneration.Biol Psychiatry. 2017 Jan 15;81(2):111-123. doi: 10.1016/j.biopsych.2016.06.025. Epub 2016 Jul 11. Biol Psychiatry. 2017. PMID: 27587263
-
The role of CREB and BDNF in neurobiology and treatment of Alzheimer's disease.Life Sci. 2020 Sep 15;257:118020. doi: 10.1016/j.lfs.2020.118020. Epub 2020 Jun 27. Life Sci. 2020. PMID: 32603820 Review.
-
Calcium-regulated signaling pathways: role in amyloid beta-induced synaptic dysfunction.Neuromolecular Med. 2004;6(1):53-64. doi: 10.1385/NMM:6:1:053. Neuromolecular Med. 2004. PMID: 15781976 Review.
Cited by
-
Comorbidity Genes of Alzheimer's Disease and Type 2 Diabetes Associated with Memory and Cognitive Function.Int J Mol Sci. 2024 Feb 12;25(4):2211. doi: 10.3390/ijms25042211. Int J Mol Sci. 2024. PMID: 38396891 Free PMC article. Review.
-
Sex Differences in Neuropathology and Cognitive Behavior in APP/PS1/tau Triple-Transgenic Mouse Model of Alzheimer's Disease.Neurosci Bull. 2018 Oct;34(5):736-746. doi: 10.1007/s12264-018-0268-9. Epub 2018 Aug 11. Neurosci Bull. 2018. PMID: 30099679 Free PMC article.
-
Short-term environmental enrichment rescues adult neurogenesis and memory deficits in APP(Sw,Ind) transgenic mice.PLoS One. 2011 Feb 9;6(2):e16832. doi: 10.1371/journal.pone.0016832. PLoS One. 2011. PMID: 21347380 Free PMC article.
-
CREB-activity and nmnat2 transcription are down-regulated prior to neurodegeneration, while NMNAT2 over-expression is neuroprotective, in a mouse model of human tauopathy.Hum Mol Genet. 2012 Jan 15;21(2):251-67. doi: 10.1093/hmg/ddr492. Epub 2011 Oct 25. Hum Mol Genet. 2012. PMID: 22027994 Free PMC article.
-
PAK4 suppresses motor neuron degeneration in hSOD1G93A -linked amyotrophic lateral sclerosis cell and rat models.Cell Prolif. 2021 Apr;54(4):e13003. doi: 10.1111/cpr.13003. Epub 2021 Feb 21. Cell Prolif. 2021. PMID: 33615605 Free PMC article.
References
-
- Belfield JL, Whittaker C, Cader MZ, Chawla S. Differential effects of Ca2+ and cAMP on transcription mediated by MEF2D and cAMP-response element-binding protein in hippocampal neurons. J Biol Chem. 2006;281:27724–27732. - PubMed
-
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous
