A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia
- PMID: 20613862
- PMCID: PMC2893954
- DOI: 10.1371/journal.pbio.1000408
A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia
Abstract
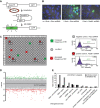
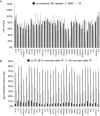
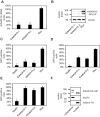
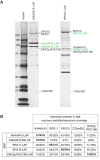
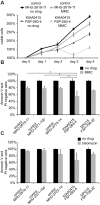
DNA repair is essential to maintain genome integrity, and genes with roles in DNA repair are frequently mutated in a variety of human diseases. Repair via homologous recombination typically restores the original DNA sequence without introducing mutations, and a number of genes that are required for homologous recombination DNA double-strand break repair (HR-DSBR) have been identified. However, a systematic analysis of this important DNA repair pathway in mammalian cells has not been reported. Here, we describe a genome-scale endoribonuclease-prepared short interfering RNA (esiRNA) screen for genes involved in DNA double strand break repair. We report 61 genes that influenced the frequency of HR-DSBR and characterize in detail one of the genes that decreased the frequency of HR-DSBR. We show that the gene KIAA0415 encodes a putative helicase that interacts with SPG11 and SPG15, two proteins mutated in hereditary spastic paraplegia (HSP). We identify mutations in HSP patients, discovering KIAA0415/SPG48 as a novel HSP-associated gene, and show that a KIAA0415/SPG48 mutant cell line is more sensitive to DNA damaging drugs. We present the first genome-scale survey of HR-DSBR in mammalian cells providing a dataset that should accelerate the discovery of novel genes with roles in DNA repair and associated medical conditions. The discovery that proteins forming a novel protein complex are required for efficient HR-DSBR and are mutated in patients suffering from HSP suggests a link between HSP and DNA repair.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Overlapping phenotypes in complex spastic paraplegias SPG11, SPG15, SPG35 and SPG48.Brain. 2014 Jul;137(Pt 7):1907-20. doi: 10.1093/brain/awu121. Epub 2014 May 15. Brain. 2014. PMID: 24833714
-
ZFYVE26/SPASTIZIN and SPG11/SPATACSIN mutations in hereditary spastic paraplegia types AR-SPG15 and AR-SPG11 have different effects on autophagy and endocytosis.Autophagy. 2019 Jan;15(1):34-57. doi: 10.1080/15548627.2018.1507438. Epub 2018 Sep 13. Autophagy. 2019. PMID: 30081747 Free PMC article.
-
Inhibiting mitochondrial fission rescues degeneration in hereditary spastic paraplegia neurons.Brain. 2022 Nov 21;145(11):4016-4031. doi: 10.1093/brain/awab488. Brain. 2022. PMID: 35026838 Free PMC article.
-
Genotype-phenotype associations in hereditary spastic paraplegia: a systematic review and meta-analysis on 13,570 patients.J Neurol. 2021 Jun;268(6):2065-2082. doi: 10.1007/s00415-019-09633-1. Epub 2019 Nov 19. J Neurol. 2021. PMID: 31745725 Review.
-
[A case of spastic paraplegia 48 with a novel mutation in the AP5Z1 gene].Rinsho Shinkeigaku. 2020 Aug 7;60(8):543-548. doi: 10.5692/clinicalneurol.60.cn-001419. Epub 2020 Jul 7. Rinsho Shinkeigaku. 2020. PMID: 32641631 Review. Japanese.
Cited by
-
Interaction between AP-5 and the hereditary spastic paraplegia proteins SPG11 and SPG15.Mol Biol Cell. 2013 Aug;24(16):2558-69. doi: 10.1091/mbc.E13-03-0170. Epub 2013 Jul 3. Mol Biol Cell. 2013. PMID: 23825025 Free PMC article.
-
A Systematic Analysis of Factors Localized to Damaged Chromatin Reveals PARP-Dependent Recruitment of Transcription Factors.Cell Rep. 2015 Jun 9;11(9):1486-500. doi: 10.1016/j.celrep.2015.04.053. Epub 2015 May 21. Cell Rep. 2015. PMID: 26004182 Free PMC article.
-
APRIN is a cell cycle specific BRCA2-interacting protein required for genome integrity and a predictor of outcome after chemotherapy in breast cancer.EMBO J. 2012 Mar 7;31(5):1160-76. doi: 10.1038/emboj.2011.490. Epub 2012 Jan 31. EMBO J. 2012. PMID: 22293751 Free PMC article.
-
Hereditary spastic paraplegia: Novel insights into the pathogenesis and management.SAGE Open Med. 2023 Dec 29;12:20503121231221941. doi: 10.1177/20503121231221941. eCollection 2024. SAGE Open Med. 2023. PMID: 38162912 Free PMC article. Review.
-
Loss of AP-5 results in accumulation of aberrant endolysosomes: defining a new type of lysosomal storage disease.Hum Mol Genet. 2015 Sep 1;24(17):4984-96. doi: 10.1093/hmg/ddv220. Epub 2015 Jun 17. Hum Mol Genet. 2015. PMID: 26085577 Free PMC article.
References
-
- Hoeijmakers J. H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. - PubMed
-
- Lombard D. B, Chua K. F, Mostoslavsky R, Franco S, Gostissa M, et al. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. - PubMed
-
- Rass U, Ahel I, West S. C. Defective DNA repair and neurodegenerative disease. Cell. 2007;130:991–1004. - PubMed
-
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. - PubMed
-
- Stewart G. S, Maser R. S, Stankovic T, Bressan D. A, Kaplan M. I, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99:577–587. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

