Conserved role of intragenic DNA methylation in regulating alternative promoters
- PMID: 20613842
- PMCID: PMC3998662
- DOI: 10.1038/nature09165
Conserved role of intragenic DNA methylation in regulating alternative promoters
Abstract
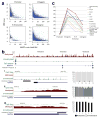
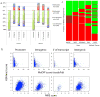
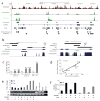
Although it is known that the methylation of DNA in 5' promoters suppresses gene expression, the role of DNA methylation in gene bodies is unclear. In mammals, tissue- and cell type-specific methylation is present in a small percentage of 5' CpG island (CGI) promoters, whereas a far greater proportion occurs across gene bodies, coinciding with highly conserved sequences. Tissue-specific intragenic methylation might reduce, or, paradoxically, enhance transcription elongation efficiency. Capped analysis of gene expression (CAGE) experiments also indicate that transcription commonly initiates within and between genes. To investigate the role of intragenic methylation, we generated a map of DNA methylation from the human brain encompassing 24.7 million of the 28 million CpG sites. From the dense, high-resolution coverage of CpG islands, the majority of methylated CpG islands were shown to be in intragenic and intergenic regions, whereas less than 3% of CpG islands in 5' promoters were methylated. The CpG islands in all three locations overlapped with RNA markers of transcription initiation, and unmethylated CpG islands also overlapped significantly with trimethylation of H3K4, a histone modification enriched at promoters. The general and CpG-island-specific patterns of methylation are conserved in mouse tissues. An in-depth investigation of the human SHANK3 locus and its mouse homologue demonstrated that this tissue-specific DNA methylation regulates intragenic promoter activity in vitro and in vivo. These methylation-regulated, alternative transcripts are expressed in a tissue- and cell type-specific manner, and are expressed differentially within a single cell type from distinct brain regions. These results support a major role for intragenic methylation in regulating cell context-specific alternative promoters in gene bodies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Comment in
-
DNA methylation: Looking beyond promoters.Nat Rev Genet. 2010 Sep;11(9):596. doi: 10.1038/nrg2851. Epub 2010 Aug 3. Nat Rev Genet. 2010. PMID: 20680023 No abstract available.
Similar articles
-
Cell type-specific DNA methylation at intragenic CpG islands in the immune system.Genome Res. 2011 Jul;21(7):1074-86. doi: 10.1101/gr.118703.110. Epub 2011 May 31. Genome Res. 2011. PMID: 21628449 Free PMC article.
-
Orphan CpG islands identify numerous conserved promoters in the mammalian genome.PLoS Genet. 2010 Sep 23;6(9):e1001134. doi: 10.1371/journal.pgen.1001134. PLoS Genet. 2010. PMID: 20885785 Free PMC article.
-
Contrasting chromatin organization of CpG islands and exons in the human genome.Genome Biol. 2010;11(7):R70. doi: 10.1186/gb-2010-11-7-r70. Epub 2010 Jul 5. Genome Biol. 2010. PMID: 20602769 Free PMC article.
-
Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation.Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194567. doi: 10.1016/j.bbagrm.2020.194567. Epub 2020 Apr 29. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32360393 Free PMC article. Review.
-
The DNA methylation paradox.Trends Genet. 1999 Jan;15(1):34-7. doi: 10.1016/s0168-9525(98)01636-9. Trends Genet. 1999. PMID: 10087932 Review.
Cited by
-
Global epigenomic reconfiguration during mammalian brain development.Science. 2013 Aug 9;341(6146):1237905. doi: 10.1126/science.1237905. Epub 2013 Jul 4. Science. 2013. PMID: 23828890 Free PMC article.
-
On the presence and role of human gene-body DNA methylation.Oncotarget. 2012 Apr;3(4):462-74. doi: 10.18632/oncotarget.497. Oncotarget. 2012. PMID: 22577155 Free PMC article.
-
MAPT Locus in Parkinson's Disease Patients of Ashkenazi Origin: A Stratified Analysis.Genes (Basel). 2023 Dec 28;15(1):46. doi: 10.3390/genes15010046. Genes (Basel). 2023. PMID: 38254936 Free PMC article.
-
Gene body methylation is conserved between plant orthologs and is of evolutionary consequence.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):1797-802. doi: 10.1073/pnas.1215380110. Epub 2013 Jan 14. Proc Natl Acad Sci U S A. 2013. PMID: 23319627 Free PMC article.
-
Methylomic trajectories across human fetal brain development.Genome Res. 2015 Mar;25(3):338-52. doi: 10.1101/gr.180273.114. Epub 2015 Feb 3. Genome Res. 2015. PMID: 25650246 Free PMC article.
References
-
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

