Central region of talin has a unique fold that binds vinculin and actin
- PMID: 20610383
- PMCID: PMC2937989
- DOI: 10.1074/jbc.M109.095455
Central region of talin has a unique fold that binds vinculin and actin
Abstract
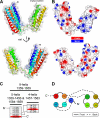
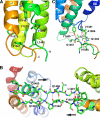
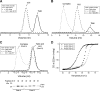
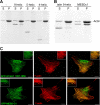
Talin is an adaptor protein that couples integrins to F-actin. Structural studies show that the N-terminal talin head contains an atypical FERM domain, whereas the N- and C-terminal parts of the talin rod include a series of α-helical bundles. However, determining the structure of the central part of the rod has proved problematic. Residues 1359-1659 are homologous to the MESDc1 gene product, and we therefore expressed this region of talin in Escherichia coli. The crystal structure shows a unique fold comprised of a 5- and 4-helix bundle. The 5-helix bundle is composed of nonsequential helices due to insertion of the 4-helix bundle into the loop at the C terminus of helix α3. The linker connecting the bundles forms a two-stranded anti-parallel β-sheet likely limiting the relative movement of the two bundles. Because the 5-helix bundle contains the N and C termini of this module, we propose that it is linked by short loops to adjacent bundles, whereas the 4-helix bundle protrudes from the rod. This suggests the 4-helix bundle has a unique role, and its pI (7.8) is higher than other rod domains. Both helical bundles contain vinculin-binding sites but that in the isolated 5-helix bundle is cryptic, whereas that in the isolated 4-helix bundle is constitutively active. In contrast, both bundles are required for actin binding. Finally, we show that the MESDc1 protein, which is predicted to have a similar fold, is a novel actin-binding protein.
Figures

Similar articles
-
The domain structure of talin: residues 1815-1973 form a five-helix bundle containing a cryptic vinculin-binding site.FEBS Lett. 2010 Jun 3;584(11):2237-41. doi: 10.1016/j.febslet.2010.04.028. Epub 2010 Apr 20. FEBS Lett. 2010. PMID: 20399778 Free PMC article.
-
The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod.J Biol Chem. 2006 Mar 17;281(11):7458-67. doi: 10.1074/jbc.M508058200. Epub 2006 Jan 9. J Biol Chem. 2006. PMID: 16407302
-
Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin.Protein Sci. 2011 Aug;20(8):1471-6. doi: 10.1002/pro.671. Protein Sci. 2011. PMID: 21648001 Free PMC article.
-
Integrin connections to the cytoskeleton through talin and vinculin.Biochem Soc Trans. 2008 Apr;36(Pt 2):235-9. doi: 10.1042/BST0360235. Biochem Soc Trans. 2008. PMID: 18363566 Review.
-
Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion.Biochem Soc Trans. 2004 Nov;32(Pt 5):831-6. doi: 10.1042/BST0320831. Biochem Soc Trans. 2004. PMID: 15494027 Review.
Cited by
-
Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics.PLoS One. 2012;7(4):e34461. doi: 10.1371/journal.pone.0034461. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496808 Free PMC article.
-
The MeshCODE to scale-visualising synaptic binary information.Front Cell Neurosci. 2022 Nov 18;16:1014629. doi: 10.3389/fncel.2022.1014629. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36467609 Free PMC article.
-
Significance of talin in cancer progression and metastasis.Int Rev Cell Mol Biol. 2011;289:117-47. doi: 10.1016/B978-0-12-386039-2.00004-3. Int Rev Cell Mol Biol. 2011. PMID: 21749900 Free PMC article. Review.
-
Two modes of integrin activation form a binary molecular switch in adhesion maturation.Mol Biol Cell. 2013 May;24(9):1354-62. doi: 10.1091/mbc.E12-09-0695. Epub 2013 Mar 6. Mol Biol Cell. 2013. PMID: 23468527 Free PMC article.
-
Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions.Elife. 2016 Jul 13;5:e18124. doi: 10.7554/eLife.18124. Elife. 2016. PMID: 27410476 Free PMC article.
References
-
- Critchley D. R., Gingras A. R. (2008) J. Cell Sci. 121, 1345–1347 - PubMed
-
- Critchley D. R. (2009) Annu. Rev. Biophys. 38, 235–254 - PubMed
-
- Calderwood D. A. (2004) J. Cell Sci. 117, 657–666 - PubMed
-
- Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., Calderwood D. A. (2003) Science 302, 103–106 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

